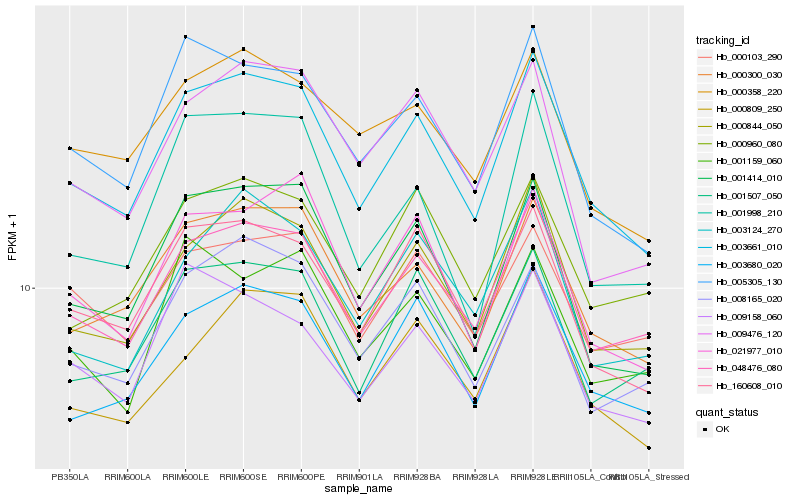
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003661_010 |
0.0 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000844_050 |
0.048972321 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 3 |
Hb_048476_080 |
0.0508351234 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 4 |
Hb_000300_030 |
0.0520191373 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 5 |
Hb_160608_010 |
0.0571874817 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 6 |
Hb_003680_020 |
0.0592829958 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_001414_010 |
0.0594250187 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 8 |
Hb_005305_130 |
0.0606624117 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 9 |
Hb_021977_010 |
0.0690348143 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 10 |
Hb_000809_250 |
0.0714740721 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 11 |
Hb_001507_050 |
0.0721998516 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000358_220 |
0.0724306945 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 13 |
Hb_009158_060 |
0.0728584821 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 14 |
Hb_008165_020 |
0.0730278895 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000103_290 |
0.0787527749 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 16 |
Hb_000960_080 |
0.0794709272 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_003124_270 |
0.0801313103 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 18 |
Hb_001998_210 |
0.0804420684 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_009476_120 |
0.0809792981 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 20 |
Hb_001159_060 |
0.0833288314 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |