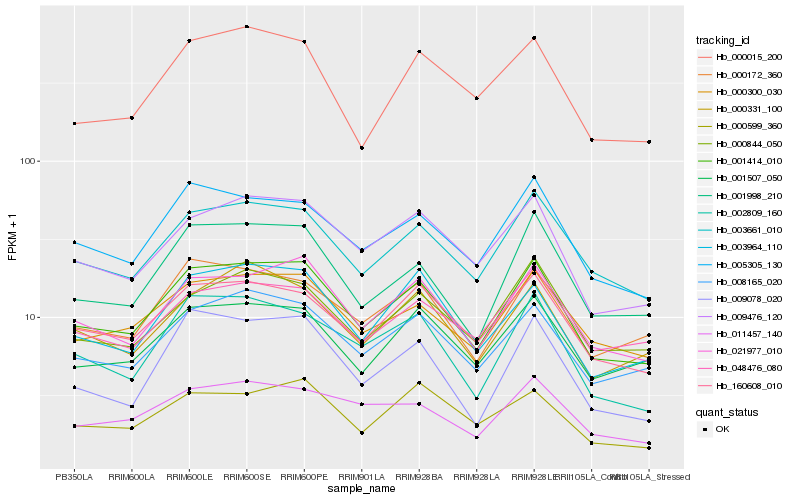
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001414_010 |
0.0 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 2 |
Hb_021977_010 |
0.0504540119 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 3 |
Hb_003661_010 |
0.0594250187 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 4 |
Hb_009476_120 |
0.0611811949 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 5 |
Hb_008165_020 |
0.0622201727 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000300_030 |
0.0634333336 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 7 |
Hb_160608_010 |
0.0659165099 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 8 |
Hb_005305_130 |
0.0659955748 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 9 |
Hb_001998_210 |
0.069763565 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_002809_160 |
0.073175077 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 11 |
Hb_003964_110 |
0.0747520376 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000172_360 |
0.0754907465 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |
| 13 |
Hb_001507_050 |
0.0769171492 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_000844_050 |
0.0793935459 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 15 |
Hb_000331_100 |
0.0808959764 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000015_200 |
0.0824392046 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: heat shock cognate 70 kDa protein 2-like [Jatropha curcas] |
| 17 |
Hb_000599_360 |
0.0834090279 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 18 |
Hb_011457_140 |
0.0838419443 |
- |
- |
PREDICTED: uncharacterized protein LOC105649947 [Jatropha curcas] |
| 19 |
Hb_048476_080 |
0.0850676792 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 20 |
Hb_009078_020 |
0.0855257228 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |