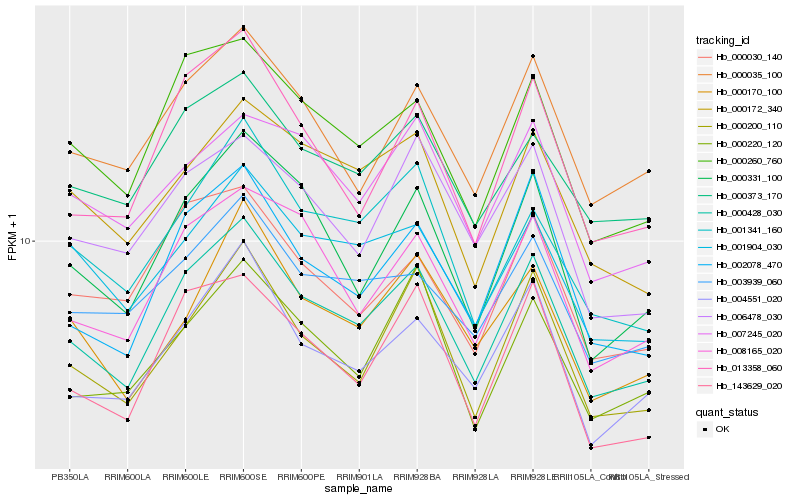
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000428_030 |
0.0 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 2 |
Hb_000260_760 |
0.0596878806 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 3 |
Hb_002078_470 |
0.0638138883 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000331_100 |
0.0640484357 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 5 |
Hb_008165_020 |
0.0656428732 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004551_020 |
0.0669221797 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 7 |
Hb_000200_110 |
0.0682813667 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 8 |
Hb_013358_060 |
0.0692205344 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 9 |
Hb_143629_020 |
0.0706800669 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 10 |
Hb_000035_100 |
0.0713390153 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_001341_160 |
0.0731947842 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000172_340 |
0.0741383793 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 13 |
Hb_000373_170 |
0.0742848185 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 14 |
Hb_001904_030 |
0.0751348266 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 15 |
Hb_003939_060 |
0.0769509358 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000220_120 |
0.0783649228 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000170_100 |
0.0796003194 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 18 |
Hb_000030_140 |
0.0799455627 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006478_030 |
0.0805271952 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 20 |
Hb_007245_020 |
0.081410072 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |