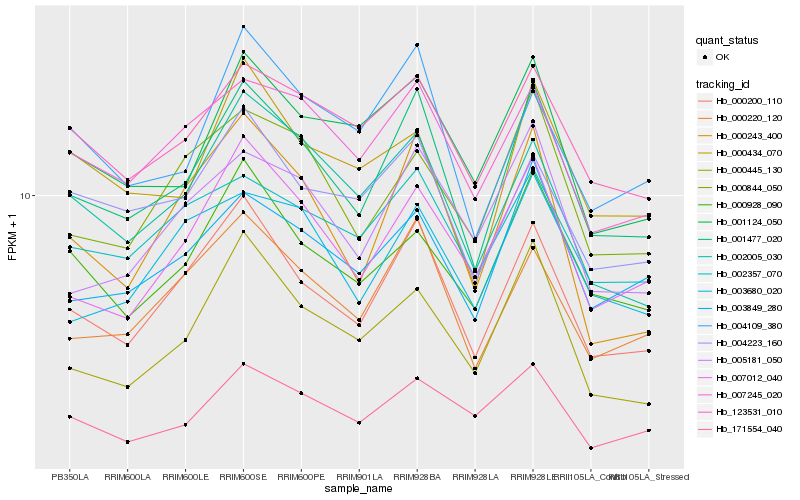
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001477_020 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105646886 isoform X3 [Jatropha curcas] |
| 2 |
Hb_000200_110 |
0.0591394239 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000220_120 |
0.0754514044 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 4 |
Hb_003849_280 |
0.0842947619 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 5 |
Hb_007012_040 |
0.0876190551 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 6 |
Hb_007245_020 |
0.0918303436 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 7 |
Hb_001124_050 |
0.0928266054 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 8 |
Hb_123531_010 |
0.0930628243 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 9 |
Hb_000928_090 |
0.0954223123 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 10 |
Hb_004109_380 |
0.095837267 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 11 |
Hb_002005_030 |
0.0968640774 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 12 |
Hb_002357_070 |
0.0969046913 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 13 |
Hb_004223_160 |
0.0970454803 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000243_400 |
0.0977229536 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 15 |
Hb_005181_050 |
0.097764587 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 16 |
Hb_000434_070 |
0.0984335014 |
- |
- |
PREDICTED: uncharacterized transmembrane protein DDB_G0289901 [Jatropha curcas] |
| 17 |
Hb_000844_050 |
0.0987912624 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 18 |
Hb_171554_040 |
0.100301222 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase [Populus euphratica] |
| 19 |
Hb_003680_020 |
0.1004324003 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000445_130 |
0.100478676 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |