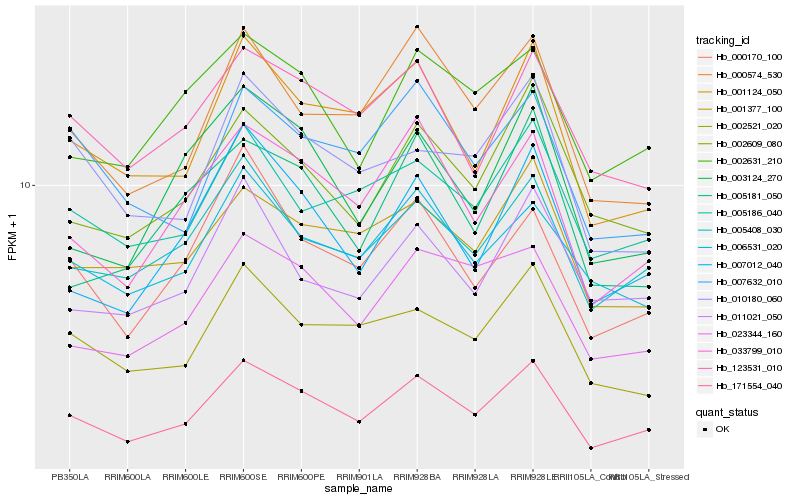
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171554_040 |
0.0 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase [Populus euphratica] |
| 2 |
Hb_033799_010 |
0.0669825422 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 3 |
Hb_001124_050 |
0.0724037501 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 4 |
Hb_000574_530 |
0.0762847666 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 5 |
Hb_007012_040 |
0.0778644659 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 6 |
Hb_005408_030 |
0.0851410042 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 7 |
Hb_000170_100 |
0.0857108925 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 8 |
Hb_002609_080 |
0.0862525538 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_002521_020 |
0.0873963492 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_023344_160 |
0.0891090882 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 11 |
Hb_003124_270 |
0.0929913142 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 12 |
Hb_005186_040 |
0.0942840418 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001377_100 |
0.0953175627 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006531_020 |
0.0960200849 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 15 |
Hb_010180_060 |
0.0969066582 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 16 |
Hb_007632_010 |
0.0971471674 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_123531_010 |
0.0973315728 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 18 |
Hb_002631_210 |
0.0985362351 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 19 |
Hb_011021_050 |
0.0991881852 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005181_050 |
0.0994580475 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |