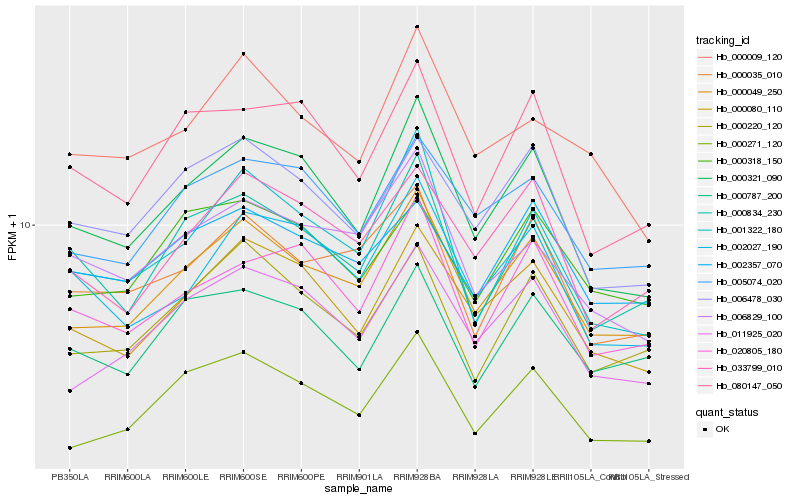
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000049_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 2 |
Hb_011925_020 |
0.0595339174 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 3 |
Hb_000321_090 |
0.0677678923 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 4 |
Hb_000080_110 |
0.0678425411 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_033799_010 |
0.0729133508 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 6 |
Hb_005074_020 |
0.0729730359 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000220_120 |
0.0732199569 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000035_010 |
0.0765037483 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001322_180 |
0.084825513 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 10 |
Hb_006478_030 |
0.0858530168 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 11 |
Hb_000834_230 |
0.0871696251 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 12 |
Hb_002357_070 |
0.0888834066 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 13 |
Hb_000318_150 |
0.0894190467 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 14 |
Hb_020805_180 |
0.0912668607 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 15 |
Hb_006829_100 |
0.0916719456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000787_200 |
0.091909237 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000009_120 |
0.0919639305 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 18 |
Hb_000271_120 |
0.0922590695 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Nelumbo nucifera] |
| 19 |
Hb_002027_190 |
0.0923919604 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 20 |
Hb_080147_050 |
0.0932486724 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |