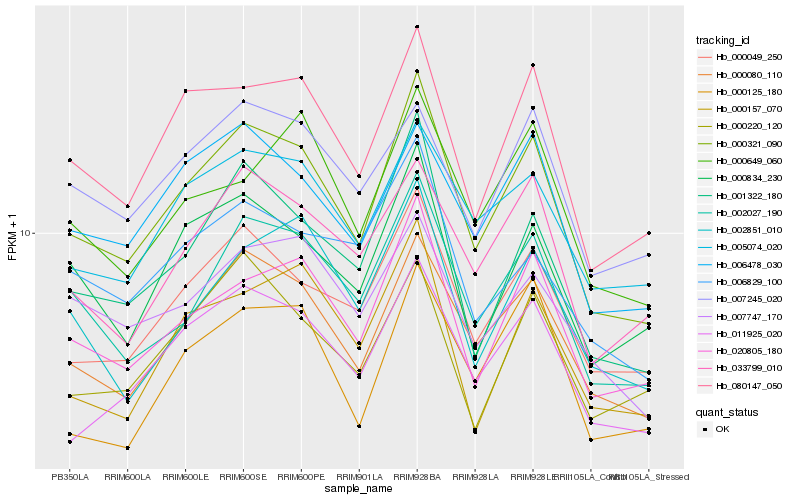
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000321_090 |
0.0 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 2 |
Hb_000080_110 |
0.0553828746 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_080147_050 |
0.0658122308 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 4 |
Hb_002027_190 |
0.0660919046 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 5 |
Hb_020805_180 |
0.0666043953 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 6 |
Hb_000049_250 |
0.0677678923 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001322_180 |
0.0731377498 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 8 |
Hb_011925_020 |
0.0753042283 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 9 |
Hb_006478_030 |
0.0807434289 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 10 |
Hb_000834_230 |
0.0820197358 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 11 |
Hb_033799_010 |
0.0835747439 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 12 |
Hb_005074_020 |
0.0840719025 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002851_010 |
0.0910128841 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 14 |
Hb_000649_060 |
0.0912694952 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 15 |
Hb_000157_070 |
0.0916075813 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 16 |
Hb_007747_170 |
0.0917213535 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 17 |
Hb_007245_020 |
0.0921313512 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 18 |
Hb_006829_100 |
0.0938611258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000220_120 |
0.0943381184 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000125_180 |
0.0945497607 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |