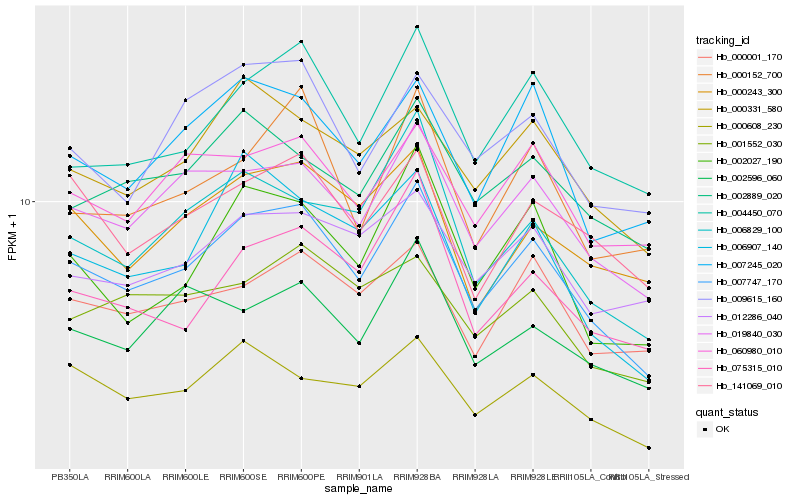
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007747_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 2 |
Hb_019840_030 |
0.0621084337 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004450_070 |
0.0798636218 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 4 |
Hb_000608_230 |
0.0805583265 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 5 |
Hb_007245_020 |
0.0823694782 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 6 |
Hb_000331_580 |
0.0842287726 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 7 |
Hb_002027_190 |
0.0842743192 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 8 |
Hb_006829_100 |
0.0848473672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000243_300 |
0.0858508112 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_006907_140 |
0.0862934428 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000001_170 |
0.0863785273 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_141069_010 |
0.0867014292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_060980_010 |
0.0868044552 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 14 |
Hb_075315_010 |
0.0882397307 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 15 |
Hb_009615_160 |
0.0882890156 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 16 |
Hb_012286_040 |
0.0890309773 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 17 |
Hb_001552_030 |
0.0891576723 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002596_060 |
0.0900578999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002889_020 |
0.090145604 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 20 |
Hb_000152_700 |
0.0904428858 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |