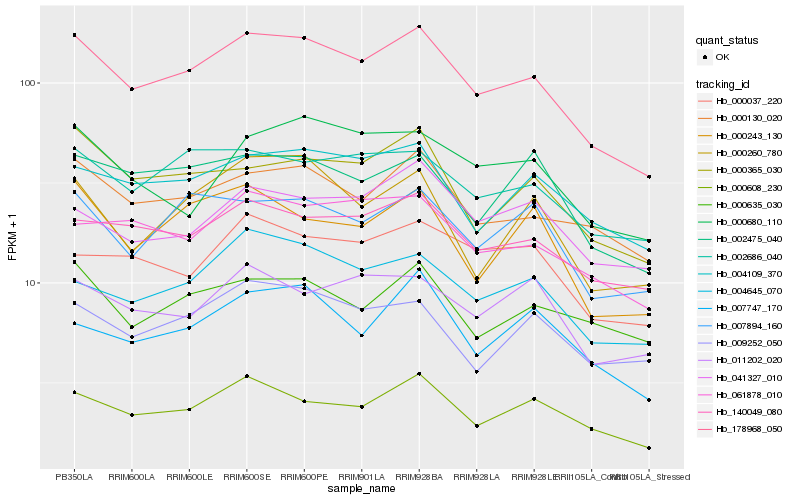
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_178968_050 |
0.0 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 2 |
Hb_000608_230 |
0.071649793 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000130_020 |
0.0718968239 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 4 |
Hb_140049_080 |
0.0775807724 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 5 |
Hb_000365_030 |
0.0790924433 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 6 |
Hb_002686_040 |
0.0791715817 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 7 |
Hb_004109_370 |
0.0794025096 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 8 |
Hb_004645_070 |
0.0807597135 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 9 |
Hb_007894_160 |
0.082256714 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 10 |
Hb_061878_010 |
0.0845969464 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 11 |
Hb_000243_130 |
0.0877242916 |
- |
- |
hypothetical protein M569_10795, partial [Genlisea aurea] |
| 12 |
Hb_002475_040 |
0.0885932963 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000260_780 |
0.0903742336 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Jatropha curcas] |
| 14 |
Hb_009252_050 |
0.0908801262 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_007747_170 |
0.0920460323 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 16 |
Hb_000635_030 |
0.0923217969 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 17 |
Hb_000037_220 |
0.0924968105 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 18 |
Hb_000680_110 |
0.0936687374 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 19 |
Hb_011202_020 |
0.0941991613 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_041327_010 |
0.0949351288 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |