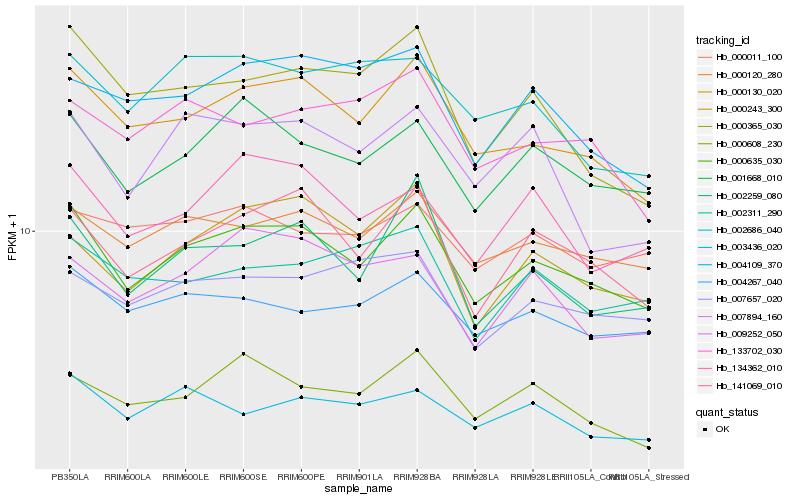
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000635_030 |
0.0 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 2 |
Hb_000130_020 |
0.0519722529 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 3 |
Hb_141069_010 |
0.0593698138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001668_010 |
0.0635285892 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 5 |
Hb_000120_280 |
0.0650826066 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 6 |
Hb_002259_080 |
0.0718902069 |
- |
- |
protein transport protein sec23, putative [Ricinus communis] |
| 7 |
Hb_004267_040 |
0.0779358763 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 8 |
Hb_134362_010 |
0.0809886084 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 9 |
Hb_000243_300 |
0.0837194921 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_003436_020 |
0.0847228941 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000608_230 |
0.0859395828 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 12 |
Hb_007657_020 |
0.0880999466 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 13 |
Hb_000365_030 |
0.088533042 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 14 |
Hb_004109_370 |
0.0890391149 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 15 |
Hb_133702_030 |
0.089449732 |
- |
- |
PREDICTED: uric acid degradation bifunctional protein TTL isoform X1 [Jatropha curcas] |
| 16 |
Hb_009252_050 |
0.089889204 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_002311_290 |
0.0907738124 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 18 |
Hb_002686_040 |
0.0912875289 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 19 |
Hb_000011_100 |
0.0915746561 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 20 |
Hb_007894_160 |
0.091719459 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |