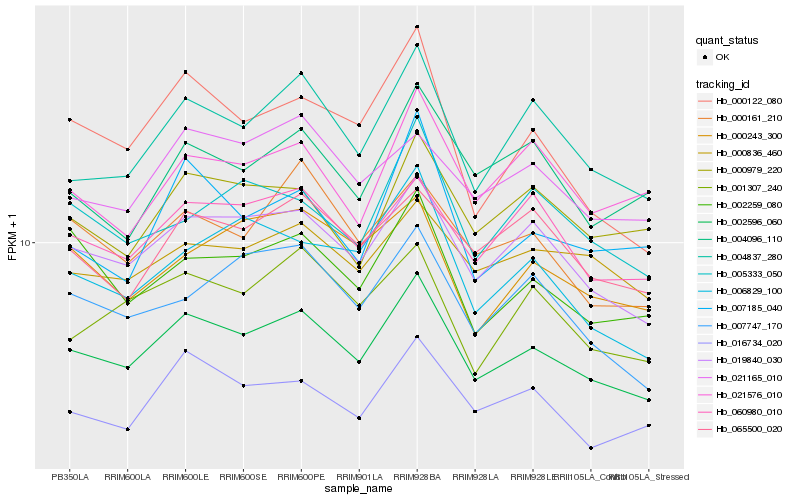
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002596_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_019840_030 |
0.0504506945 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004837_280 |
0.0719275266 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 4 |
Hb_000122_080 |
0.0802885628 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 5 |
Hb_060980_010 |
0.0809480969 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 6 |
Hb_002259_080 |
0.0833365136 |
- |
- |
protein transport protein sec23, putative [Ricinus communis] |
| 7 |
Hb_000161_210 |
0.087859919 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 8 |
Hb_001307_240 |
0.0881088409 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_016734_020 |
0.0884401672 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 10 |
Hb_000979_220 |
0.0893167148 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007747_170 |
0.0900578999 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 12 |
Hb_000836_460 |
0.0902534364 |
- |
- |
PREDICTED: uncharacterized protein LOC105628435 [Jatropha curcas] |
| 13 |
Hb_065500_020 |
0.0904995015 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 14 |
Hb_006829_100 |
0.0917683282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_021165_010 |
0.0922193597 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_005333_050 |
0.0938830993 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000243_300 |
0.0940218172 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_004096_110 |
0.0946325478 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 19 |
Hb_007185_040 |
0.0952007759 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 20 |
Hb_021576_010 |
0.0955224885 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |