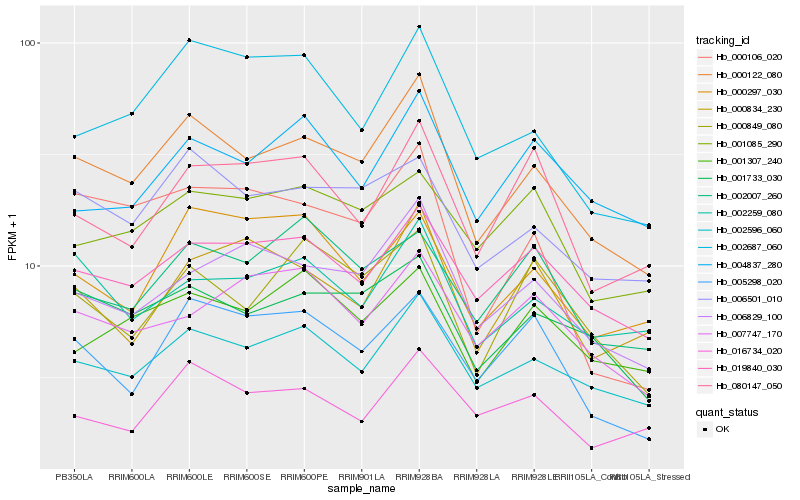
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000122_080 |
0.0 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 2 |
Hb_002596_060 |
0.0802885628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001733_030 |
0.0905192196 |
- |
- |
PREDICTED: uncharacterized protein LOC100261386 isoform X2 [Vitis vinifera] |
| 4 |
Hb_006501_010 |
0.0906364231 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3A isoform X1 [Populus euphratica] |
| 5 |
Hb_006829_100 |
0.0943364813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_019840_030 |
0.100316349 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002259_080 |
0.1093298476 |
- |
- |
protein transport protein sec23, putative [Ricinus communis] |
| 8 |
Hb_002687_060 |
0.1109349128 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 9 |
Hb_000849_080 |
0.1129981131 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005298_020 |
0.1140051965 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 2 [Jatropha curcas] |
| 11 |
Hb_016734_020 |
0.116974856 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 12 |
Hb_001307_240 |
0.1172962407 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000106_020 |
0.1178053742 |
- |
- |
PREDICTED: protein TOPLESS isoform X1 [Jatropha curcas] |
| 14 |
Hb_004837_280 |
0.1212612592 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 15 |
Hb_000834_230 |
0.1212687121 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 16 |
Hb_000297_030 |
0.1226278271 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_080147_050 |
0.1241004291 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 18 |
Hb_001085_290 |
0.1245909717 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 19 |
Hb_002007_260 |
0.1254003307 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 20 |
Hb_007747_170 |
0.1258336765 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |