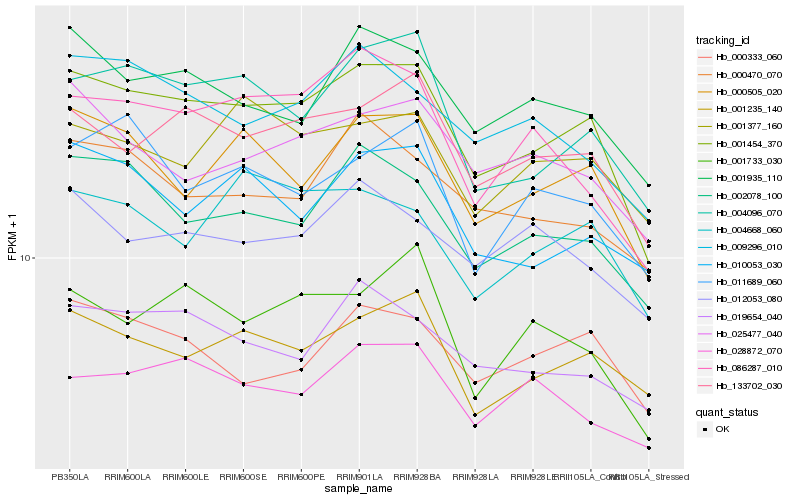
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001454_370 |
0.0 |
- |
- |
PREDICTED: glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a [Jatropha curcas] |
| 2 |
Hb_133702_030 |
0.066381005 |
- |
- |
PREDICTED: uric acid degradation bifunctional protein TTL isoform X1 [Jatropha curcas] |
| 3 |
Hb_000505_020 |
0.070242751 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 4 |
Hb_004096_070 |
0.0712115274 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_002078_100 |
0.0732894522 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 6 |
Hb_001235_140 |
0.0765174037 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 7 |
Hb_001935_110 |
0.0788975308 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 8 |
Hb_019654_040 |
0.0827530112 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 9 |
Hb_086287_010 |
0.0833631129 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_009296_010 |
0.0839026546 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 11 |
Hb_010053_030 |
0.0858412989 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 12 |
Hb_000470_070 |
0.0868230263 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 13 |
Hb_001733_030 |
0.087101159 |
- |
- |
PREDICTED: uncharacterized protein LOC100261386 isoform X2 [Vitis vinifera] |
| 14 |
Hb_000333_060 |
0.0876470637 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 15 |
Hb_025477_040 |
0.087858896 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 16 |
Hb_004668_060 |
0.0887515229 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 17 |
Hb_011689_060 |
0.0892015426 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 18 |
Hb_028872_070 |
0.0897104838 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001377_160 |
0.0908661361 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 20 |
Hb_012053_080 |
0.0910250888 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |