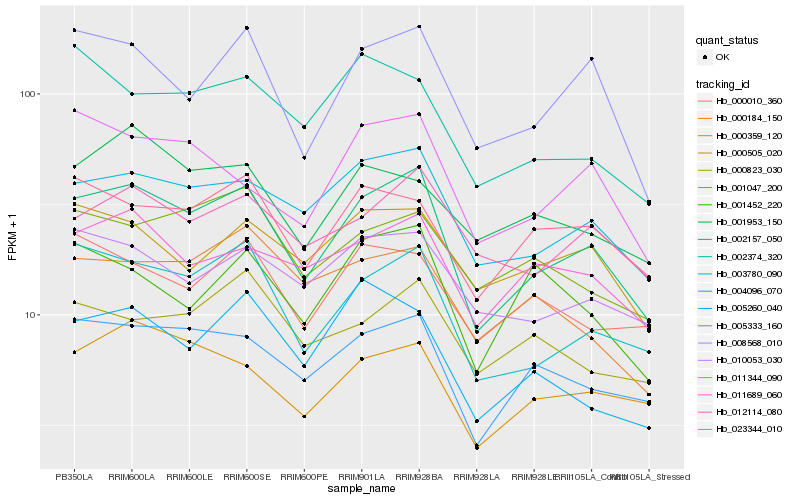
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002157_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 2 |
Hb_003780_090 |
0.0764956583 |
- |
- |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 3 |
Hb_004096_070 |
0.0808442041 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_005333_160 |
0.0882361298 |
- |
- |
hypothetical protein B456_009G230200 [Gossypium raimondii] |
| 5 |
Hb_008568_010 |
0.090512573 |
- |
- |
Phosphoenolpyruvate carboxykinase [ATP], putative [Ricinus communis] |
| 6 |
Hb_000359_120 |
0.1030070442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_011689_060 |
0.1033668683 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 8 |
Hb_023344_010 |
0.1060586733 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 9 |
Hb_012114_080 |
0.1071993358 |
- |
- |
gamma-glutamylcysteine synthetase [Hevea brasiliensis] |
| 10 |
Hb_010053_030 |
0.1076559729 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 11 |
Hb_011344_090 |
0.1124671142 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 12 |
Hb_000505_020 |
0.112943644 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 13 |
Hb_001047_200 |
0.1132096402 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 14 |
Hb_000010_360 |
0.1142838812 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 15 |
Hb_000184_150 |
0.115438837 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 16 |
Hb_005260_040 |
0.1155523063 |
- |
- |
PREDICTED: uncharacterized protein LOC105633768 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002374_320 |
0.116520159 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 18 |
Hb_000823_030 |
0.1178195233 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 19 |
Hb_001452_220 |
0.1180718501 |
- |
- |
PREDICTED: molybdopterin biosynthesis protein CNX1 [Jatropha curcas] |
| 20 |
Hb_001953_150 |
0.1180923682 |
- |
- |
PREDICTED: protein DEK isoform X2 [Jatropha curcas] |