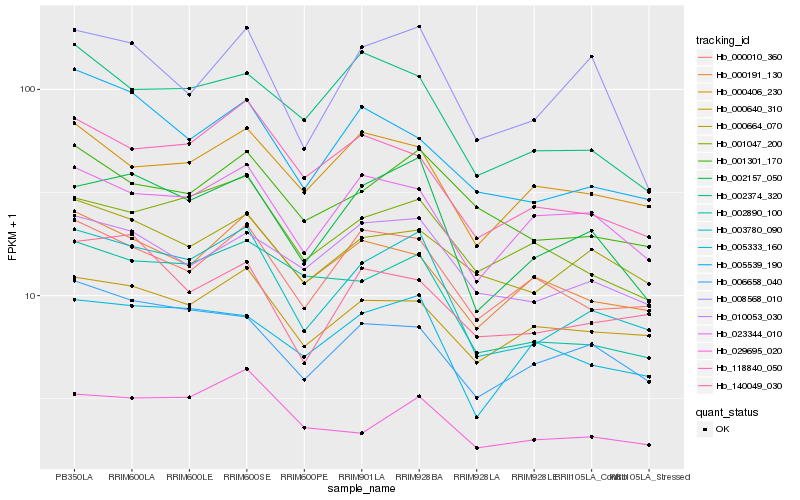
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003780_090 |
0.0 |
- |
- |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 2 |
Hb_002157_050 |
0.0764956583 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 3 |
Hb_002890_100 |
0.1021835823 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein MAD1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_118840_050 |
0.1039914443 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 5 |
Hb_029695_020 |
0.1040695531 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001047_200 |
0.1049968817 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 7 |
Hb_000010_360 |
0.105006328 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 8 |
Hb_006658_040 |
0.1054147893 |
- |
- |
hypothetical protein POPTR_0006s19640g [Populus trichocarpa] |
| 9 |
Hb_005333_160 |
0.106362453 |
- |
- |
hypothetical protein B456_009G230200 [Gossypium raimondii] |
| 10 |
Hb_001301_170 |
0.1065824889 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 11 |
Hb_000640_310 |
0.1073594004 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 12 |
Hb_005539_190 |
0.1090354857 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 13 |
Hb_010053_030 |
0.1098530588 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 14 |
Hb_000664_070 |
0.1098620349 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 15 |
Hb_000191_130 |
0.1114489494 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 16 |
Hb_002374_320 |
0.1116471809 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 17 |
Hb_023344_010 |
0.1117445218 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 18 |
Hb_140049_030 |
0.1135675171 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 19 |
Hb_008568_010 |
0.1136576432 |
- |
- |
Phosphoenolpyruvate carboxykinase [ATP], putative [Ricinus communis] |
| 20 |
Hb_000406_230 |
0.1137528991 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |