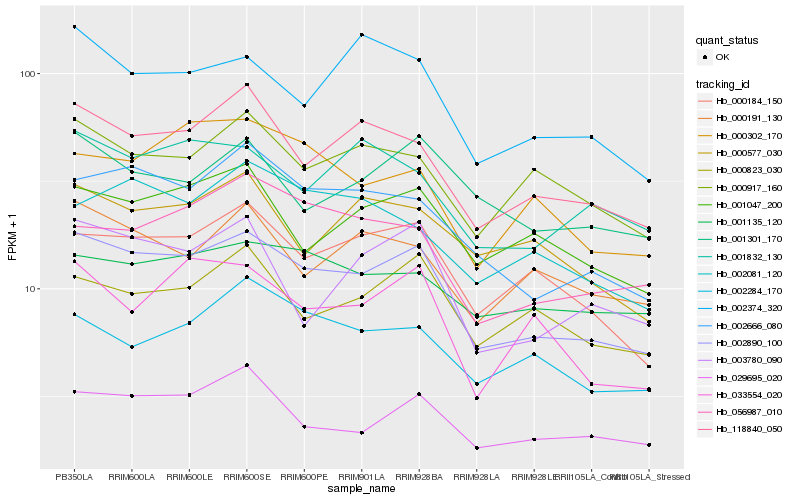
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002890_100 |
0.0 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein MAD1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_118840_050 |
0.0808468925 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 3 |
Hb_002666_080 |
0.0821632962 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_000184_150 |
0.0866863071 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 5 |
Hb_029695_020 |
0.0924343656 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001047_200 |
0.096429454 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 7 |
Hb_000917_160 |
0.0994167718 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002374_320 |
0.1011533707 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 9 |
Hb_003780_090 |
0.1021835823 |
- |
- |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 10 |
Hb_033554_020 |
0.1031080723 |
transcription factor |
TF Family: BES1 |
PREDICTED: beta-amylase 8 [Jatropha curcas] |
| 11 |
Hb_056987_010 |
0.1031193892 |
- |
- |
DNA-damage-repair/toleration protein DRT111, chloroplastic [Glycine soja] |
| 12 |
Hb_000823_030 |
0.1035985554 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 13 |
Hb_001832_130 |
0.1040978522 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 14 |
Hb_001135_120 |
0.1041608943 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000191_130 |
0.1047798296 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 16 |
Hb_000302_170 |
0.1052646929 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 17 |
Hb_002284_170 |
0.1053454405 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 18 |
Hb_000577_030 |
0.1064326197 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |
| 19 |
Hb_001301_170 |
0.1066814583 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 20 |
Hb_002081_120 |
0.1070037281 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |