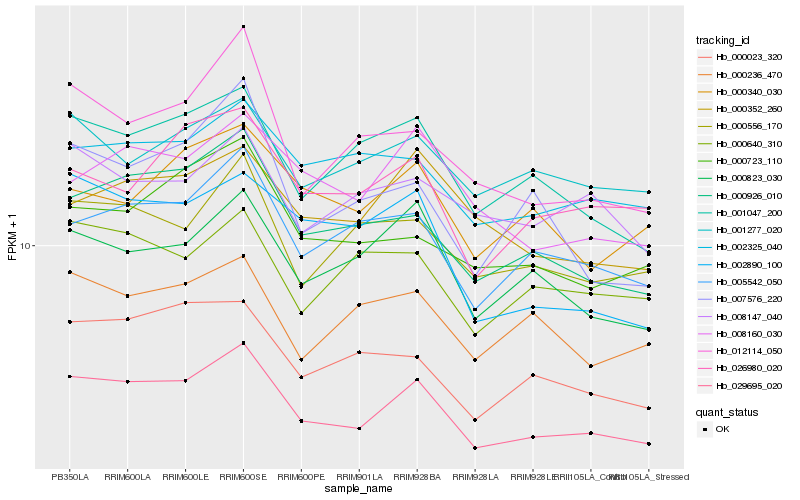
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029695_020 |
0.0 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000926_010 |
0.0694034582 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 3 |
Hb_005542_050 |
0.078687271 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 4 |
Hb_001047_200 |
0.0789109397 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 5 |
Hb_000823_030 |
0.0838030984 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 6 |
Hb_012114_050 |
0.0861807137 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000556_170 |
0.0863475899 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000023_320 |
0.0868230843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002325_040 |
0.0891287149 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000723_110 |
0.0892591609 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 11 |
Hb_008160_030 |
0.0911696913 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_002890_100 |
0.0924343656 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein MAD1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_026980_020 |
0.0929863664 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 14 |
Hb_000352_260 |
0.0946780971 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 15 |
Hb_000236_470 |
0.0953613197 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 16 |
Hb_000640_310 |
0.0956049514 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 17 |
Hb_001277_020 |
0.0959816545 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 18 |
Hb_007576_220 |
0.0986977961 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 19 |
Hb_008147_040 |
0.0991327097 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |
| 20 |
Hb_000340_030 |
0.1004950396 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |