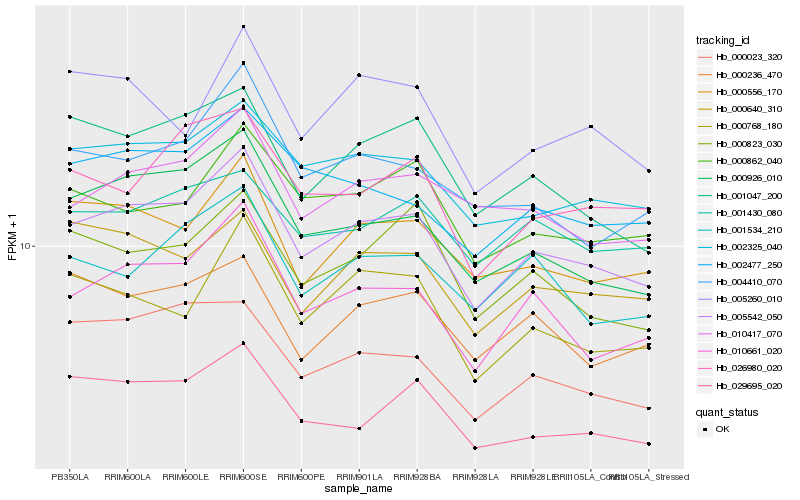
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005542_050 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 2 |
Hb_010661_020 |
0.0539463911 |
- |
- |
PREDICTED: uncharacterized protein LOC105630767 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002325_040 |
0.0603813991 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000926_010 |
0.0604351758 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 5 |
Hb_000556_170 |
0.0670626144 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 6 |
Hb_010417_070 |
0.0705693469 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002477_250 |
0.0735518352 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 8 |
Hb_001534_210 |
0.0736986572 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001047_200 |
0.0740009504 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 10 |
Hb_004410_070 |
0.0763820472 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 11 |
Hb_000640_310 |
0.0776686804 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 12 |
Hb_000768_180 |
0.0779849074 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 13 |
Hb_026980_020 |
0.0780974573 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 14 |
Hb_029695_020 |
0.078687271 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001430_080 |
0.0792357756 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 16 |
Hb_000862_040 |
0.0796320473 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 17 |
Hb_000023_320 |
0.0796800263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000823_030 |
0.0801533377 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 19 |
Hb_000236_470 |
0.0807383186 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 20 |
Hb_005260_010 |
0.0829049107 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |