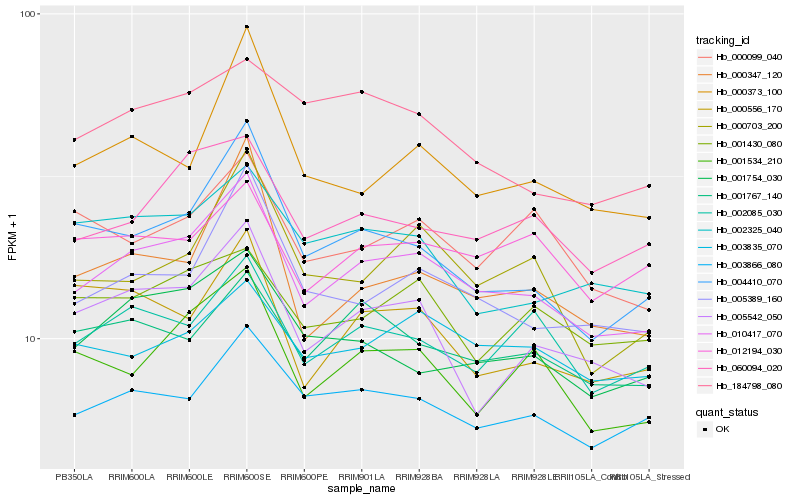
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010417_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005389_160 |
0.0626765305 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 3 |
Hb_060094_020 |
0.0682698163 |
desease resistance |
Gene Name: ATP_bind_1 |
xpa-binding protein, putative [Ricinus communis] |
| 4 |
Hb_003866_080 |
0.0685918364 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 5 |
Hb_001534_210 |
0.0690447276 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004410_070 |
0.069860363 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 7 |
Hb_002085_030 |
0.069887429 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 8 |
Hb_005542_050 |
0.0705693469 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 9 |
Hb_001754_030 |
0.071262177 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 10 |
Hb_001767_140 |
0.0732806276 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 11 |
Hb_000347_120 |
0.0739359848 |
- |
- |
PREDICTED: uncharacterized protein LOC105631363 [Jatropha curcas] |
| 12 |
Hb_000703_200 |
0.0772915709 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_003835_070 |
0.0773427828 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000373_100 |
0.0781182066 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 15 |
Hb_001430_080 |
0.0802103599 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 16 |
Hb_012194_030 |
0.0803766917 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002325_040 |
0.0803924868 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000556_170 |
0.0806320594 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000099_040 |
0.0809112605 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_184798_080 |
0.0812688899 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |