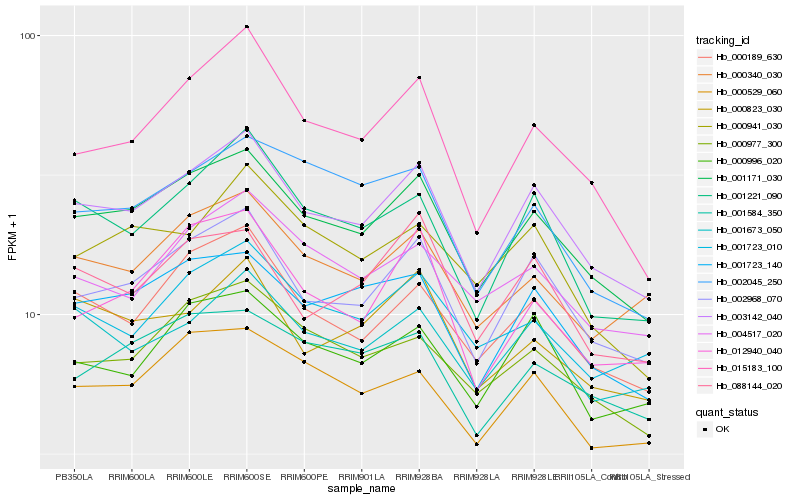
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001171_030 |
0.0 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 2 |
Hb_003142_040 |
0.0323759933 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 3 |
Hb_002968_070 |
0.0465915017 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 4 |
Hb_001723_140 |
0.0497508664 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 5 |
Hb_000189_630 |
0.0565264626 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 6 |
Hb_000977_300 |
0.05788473 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 7 |
Hb_000529_060 |
0.0589643721 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001584_350 |
0.0608653646 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 9 |
Hb_015183_100 |
0.0701804542 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 10 |
Hb_001221_090 |
0.0702516821 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000340_030 |
0.0710702612 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 12 |
Hb_002045_250 |
0.0722090643 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 13 |
Hb_001723_010 |
0.0729032659 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 14 |
Hb_004517_020 |
0.0730079242 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 15 |
Hb_000823_030 |
0.0730261854 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 16 |
Hb_012940_040 |
0.0736986195 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 17 |
Hb_001673_050 |
0.0737563704 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000996_020 |
0.0745618916 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 19 |
Hb_088144_020 |
0.0746875144 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000941_030 |
0.074945294 |
- |
- |
PREDICTED: uncharacterized protein LOC105646531 [Jatropha curcas] |