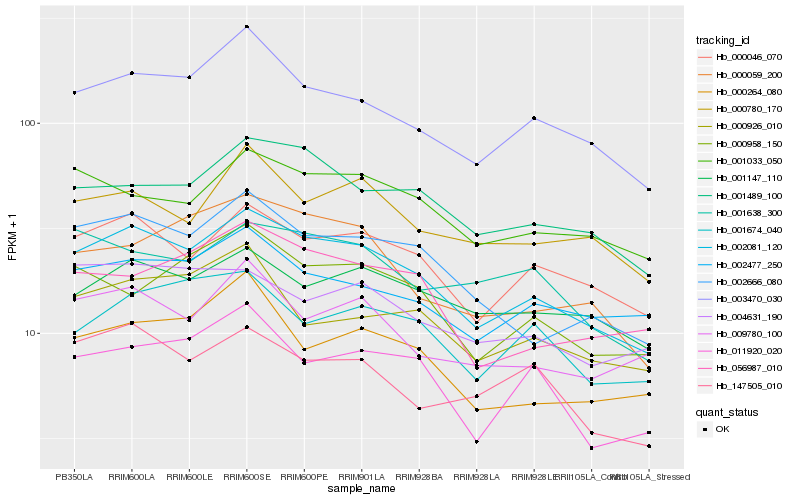
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002081_120 |
0.0 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 2 |
Hb_000046_070 |
0.0660832571 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 3 |
Hb_003470_030 |
0.0688684943 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 4 |
Hb_002666_080 |
0.0751871651 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_001489_100 |
0.0796434233 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 6 |
Hb_001033_050 |
0.0880456051 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 7 |
Hb_002477_250 |
0.0883867169 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 8 |
Hb_000958_150 |
0.0886225752 |
- |
- |
Actin, putative [Ricinus communis] |
| 9 |
Hb_001147_110 |
0.0898114211 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 10 |
Hb_009780_100 |
0.089890932 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 11 |
Hb_000780_170 |
0.0906466232 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 12 |
Hb_000059_200 |
0.0907434914 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 13 |
Hb_056987_010 |
0.0913250814 |
- |
- |
DNA-damage-repair/toleration protein DRT111, chloroplastic [Glycine soja] |
| 14 |
Hb_011920_020 |
0.0916325582 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 15 |
Hb_000264_080 |
0.0924244114 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 16 |
Hb_001674_040 |
0.0934247728 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 23 [Jatropha curcas] |
| 17 |
Hb_147505_010 |
0.094431477 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 18 |
Hb_000926_010 |
0.0951325217 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 19 |
Hb_004631_190 |
0.0952338332 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 20 |
Hb_001638_300 |
0.0956665834 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 4 [Jatropha curcas] |