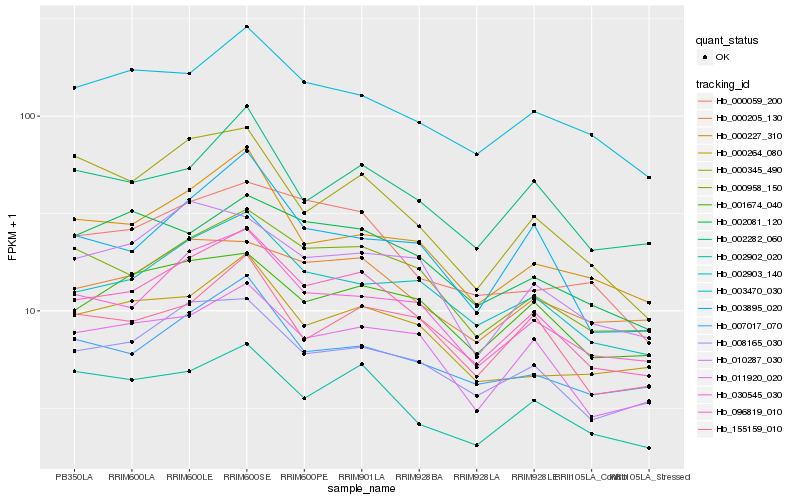
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_096819_010 |
0.0 |
- |
- |
PREDICTED: GRIP and coiled-coil domain-containing protein 2 isoform X4 [Jatropha curcas] |
| 2 |
Hb_030545_030 |
0.0639212478 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 3 |
Hb_002903_140 |
0.0693618044 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 4 |
Hb_008165_030 |
0.0710514205 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 5 |
Hb_000958_150 |
0.0718261241 |
- |
- |
Actin, putative [Ricinus communis] |
| 6 |
Hb_011920_020 |
0.0748749339 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 7 |
Hb_002902_020 |
0.08163573 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 8 |
Hb_003470_030 |
0.0837026567 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 9 |
Hb_002282_060 |
0.0849186932 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_003895_020 |
0.0852149107 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 11 |
Hb_007017_070 |
0.085522783 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000227_310 |
0.0860680192 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001674_040 |
0.0866654449 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 23 [Jatropha curcas] |
| 14 |
Hb_155159_010 |
0.0876936373 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 15 |
Hb_000059_200 |
0.0917188502 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 16 |
Hb_000264_080 |
0.0917662913 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 17 |
Hb_000205_130 |
0.0931890733 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_010287_030 |
0.095694447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002081_120 |
0.0986097924 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 20 |
Hb_000345_490 |
0.0994580005 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |