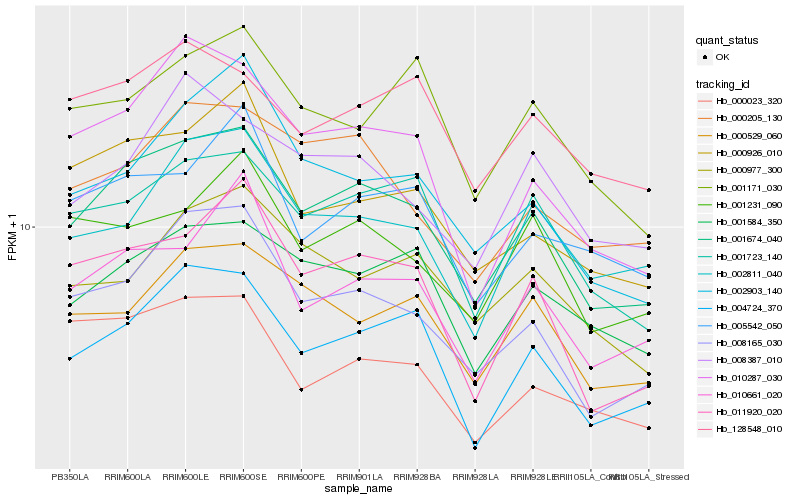
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001674_040 |
0.0 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 23 [Jatropha curcas] |
| 2 |
Hb_004724_370 |
0.0607999772 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 3 |
Hb_008165_030 |
0.0629273237 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 4 |
Hb_001723_140 |
0.0676210236 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 5 |
Hb_011920_020 |
0.069214467 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 6 |
Hb_001584_350 |
0.0715259864 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 7 |
Hb_010661_020 |
0.0727194129 |
- |
- |
PREDICTED: uncharacterized protein LOC105630767 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000529_060 |
0.0756538036 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000926_010 |
0.0758925573 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 10 |
Hb_010287_030 |
0.0760153129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000205_130 |
0.0772952196 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_001231_090 |
0.0784541732 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002903_140 |
0.0803917819 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 14 |
Hb_002811_040 |
0.0810191715 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |
| 15 |
Hb_128548_010 |
0.0826257262 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_008387_010 |
0.082927938 |
- |
- |
SMAD/FHA domain-containing protein isoform 3 [Theobroma cacao] |
| 17 |
Hb_005542_050 |
0.0831076932 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 18 |
Hb_000023_320 |
0.0844060642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001171_030 |
0.0853400806 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 20 |
Hb_000977_300 |
0.0857140207 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |