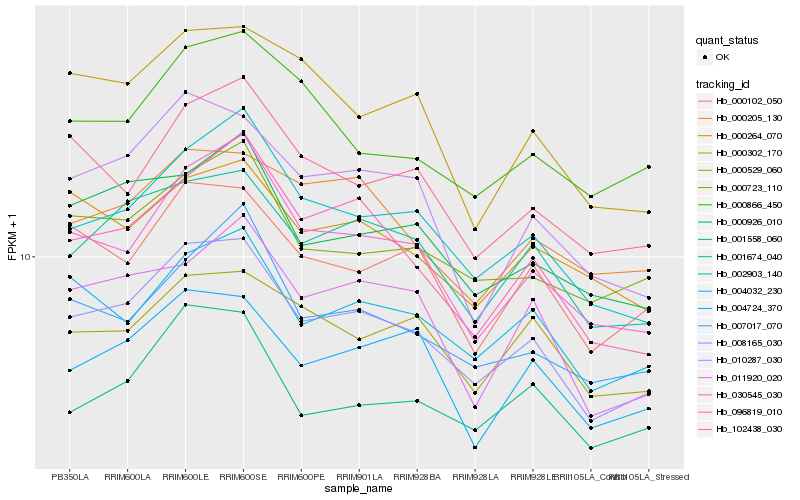
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008165_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 2 |
Hb_002903_140 |
0.0613581742 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 3 |
Hb_030545_030 |
0.0621995283 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 4 |
Hb_001674_040 |
0.0629273237 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 23 [Jatropha curcas] |
| 5 |
Hb_010287_030 |
0.0707884622 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_096819_010 |
0.0710514205 |
- |
- |
PREDICTED: GRIP and coiled-coil domain-containing protein 2 isoform X4 [Jatropha curcas] |
| 7 |
Hb_004032_230 |
0.0729002303 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 8 |
Hb_000102_050 |
0.0747523285 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000926_010 |
0.0773406263 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 10 |
Hb_000723_110 |
0.0782146514 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 11 |
Hb_007017_070 |
0.0784243472 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004724_370 |
0.0788658411 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 13 |
Hb_102438_030 |
0.0796120044 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_011920_020 |
0.0813482813 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 15 |
Hb_001558_060 |
0.0816303791 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000302_170 |
0.0828338952 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 17 |
Hb_000529_060 |
0.0862943556 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000866_450 |
0.0871290959 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 19 |
Hb_000264_070 |
0.0875161595 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 20 |
Hb_000205_130 |
0.0880597906 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |