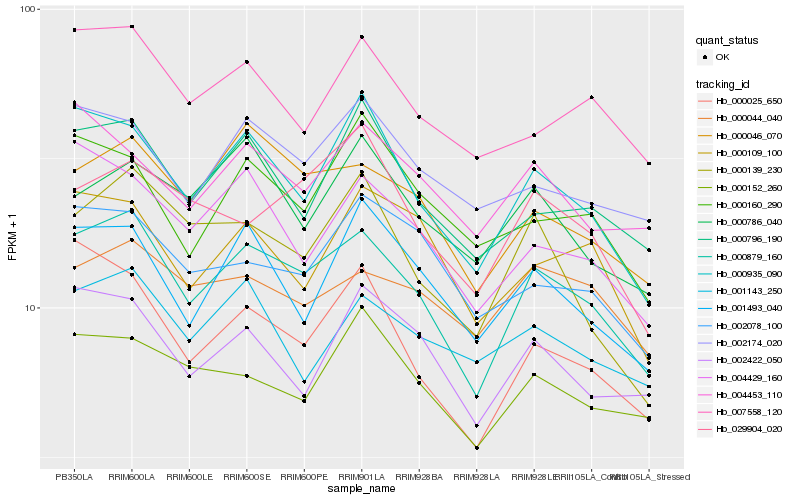
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000879_160 |
0.0 |
- |
- |
PREDICTED: F-box protein At3g12350 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000935_090 |
0.0725815559 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 3 |
Hb_000046_070 |
0.0751919093 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 4 |
Hb_000796_190 |
0.0861496038 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 5 |
Hb_004429_160 |
0.088057777 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000025_650 |
0.0882026472 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 7 |
Hb_007558_120 |
0.0910709644 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 8 |
Hb_001493_040 |
0.0911536679 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 9 |
Hb_002174_020 |
0.0915262788 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 10 |
Hb_002422_050 |
0.0933221256 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000152_260 |
0.0951375802 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 12 |
Hb_000109_100 |
0.0967559198 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Jatropha curcas] |
| 13 |
Hb_000044_040 |
0.0973791465 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 14 |
Hb_002078_100 |
0.0975542672 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 15 |
Hb_000139_230 |
0.0978583898 |
transcription factor |
TF Family: SNF2 |
PREDICTED: probable ATP-dependent DNA helicase CHR12 [Jatropha curcas] |
| 16 |
Hb_029904_020 |
0.0982617608 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 17 |
Hb_004453_110 |
0.0988649239 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 18 |
Hb_000160_290 |
0.0997040464 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_001143_250 |
0.0998686187 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 20 |
Hb_000786_040 |
0.1004057512 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 17 [Jatropha curcas] |