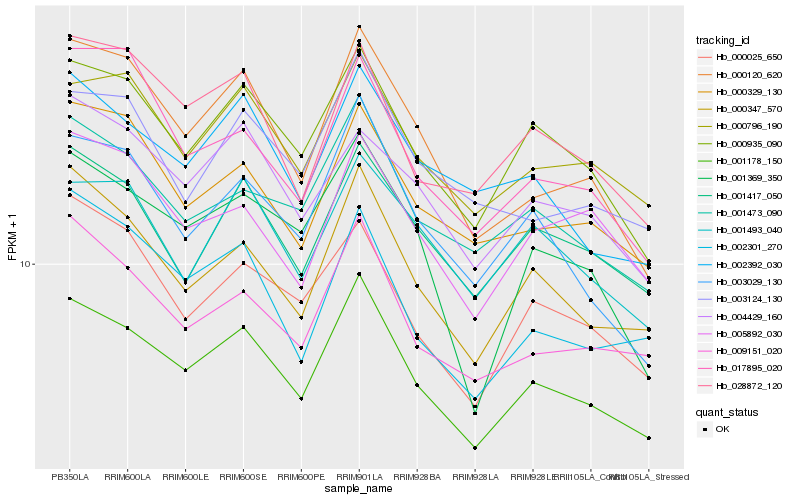
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001178_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635484 [Jatropha curcas] |
| 2 |
Hb_000935_090 |
0.0529237852 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 3 |
Hb_000347_570 |
0.0551107487 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 4 |
Hb_000796_190 |
0.0641782371 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 5 |
Hb_001473_090 |
0.0654859264 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 6 |
Hb_009151_020 |
0.0663212232 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 7 |
Hb_000120_620 |
0.0685137258 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 8 |
Hb_001417_050 |
0.07157892 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 9 |
Hb_000329_130 |
0.0722530576 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 10 |
Hb_003029_130 |
0.0729205432 |
- |
- |
carboxypeptidase regulatory region-containingprotein, putative [Ricinus communis] |
| 11 |
Hb_028872_120 |
0.0734682772 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 12 |
Hb_000025_650 |
0.0750252152 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 13 |
Hb_002392_030 |
0.0761074327 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 14 |
Hb_004429_160 |
0.077924821 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001369_350 |
0.0785124988 |
- |
- |
Cisplatin resistance-associated overexpressed protein, putative [Ricinus communis] |
| 16 |
Hb_003124_130 |
0.0794401456 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002301_270 |
0.0796172531 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 18 |
Hb_001493_040 |
0.0818461303 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 19 |
Hb_005892_030 |
0.0818644068 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 20 |
Hb_017895_020 |
0.0847100982 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |