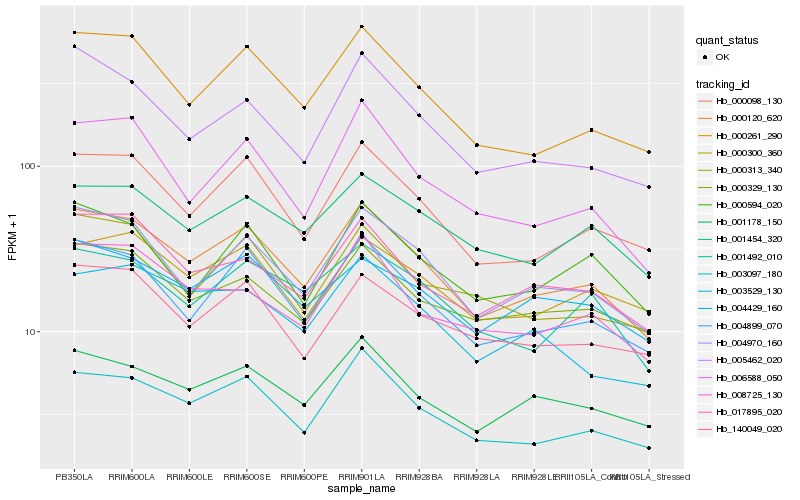
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_620 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 2 |
Hb_004899_070 |
0.0558080191 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 3 |
Hb_004970_160 |
0.0566685473 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000098_130 |
0.0581338839 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 5 |
Hb_003097_180 |
0.0589180194 |
- |
- |
PREDICTED: crossover junction endonuclease MUS81 [Jatropha curcas] |
| 6 |
Hb_000261_290 |
0.059947584 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 7 |
Hb_001178_150 |
0.0685137258 |
- |
- |
PREDICTED: uncharacterized protein LOC105635484 [Jatropha curcas] |
| 8 |
Hb_017895_020 |
0.0780031883 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 9 |
Hb_006588_050 |
0.078643606 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 10 |
Hb_000329_130 |
0.0794302064 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 11 |
Hb_001492_010 |
0.0814627963 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like isoform X2 [Populus euphratica] |
| 12 |
Hb_140049_020 |
0.081661759 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 13 |
Hb_005462_020 |
0.0823524678 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 14 |
Hb_008725_130 |
0.0830272395 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 15 |
Hb_001454_320 |
0.0844134055 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000313_340 |
0.0845673292 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 17 |
Hb_000594_020 |
0.0853827582 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004429_160 |
0.0857273595 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003529_130 |
0.0863890716 |
- |
- |
PREDICTED: ribosome biogenesis protein BOP1 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_000300_360 |
0.0864667428 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |