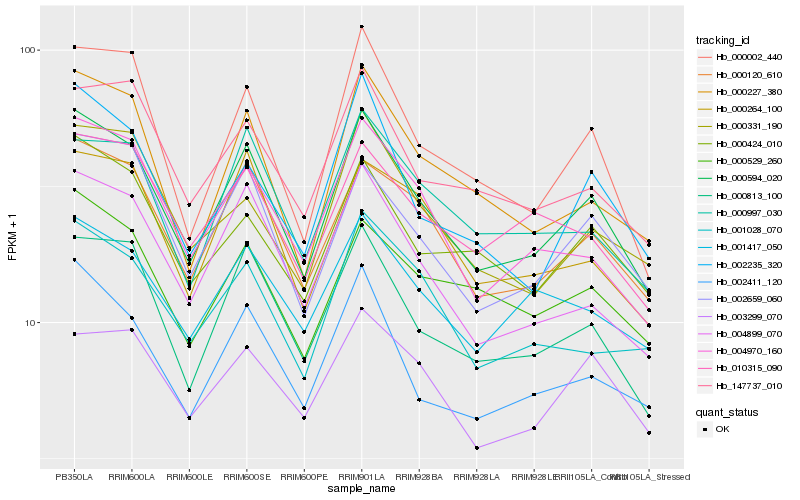
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000594_020 |
0.0 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000120_610 |
0.0530198454 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 3 |
Hb_000813_100 |
0.0579803179 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002659_060 |
0.0633578515 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 5 |
Hb_004899_070 |
0.0678372251 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 6 |
Hb_002411_120 |
0.0702008782 |
- |
- |
PREDICTED: uncharacterized protein LOC105631645 [Jatropha curcas] |
| 7 |
Hb_004970_160 |
0.0713961271 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000002_440 |
0.0727966186 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 9 |
Hb_000227_380 |
0.074518545 |
- |
- |
PREDICTED: protein TIME FOR COFFEE isoform X3 [Jatropha curcas] |
| 10 |
Hb_000264_100 |
0.0753305908 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_147737_010 |
0.076142236 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000997_030 |
0.0782914012 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 13 |
Hb_000424_010 |
0.078686566 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000529_260 |
0.0788537527 |
- |
- |
PREDICTED: protein SGT1 homolog At5g65490 [Jatropha curcas] |
| 15 |
Hb_002235_320 |
0.0793167328 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001417_050 |
0.0795499184 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 17 |
Hb_010315_090 |
0.0816545675 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 18 |
Hb_001028_070 |
0.0824412802 |
- |
- |
PREDICTED: WD repeat-containing protein 75 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000331_190 |
0.0829032739 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 20 |
Hb_003299_070 |
0.0829574374 |
- |
- |
AMP deaminase [Theobroma cacao] |