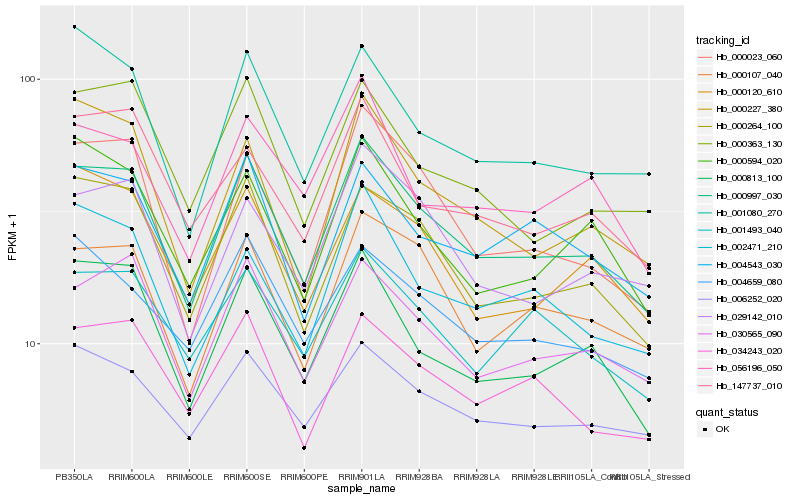
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000997_030 |
0.0 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 2 |
Hb_000264_100 |
0.0574265919 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_030565_090 |
0.0576529347 |
- |
- |
PREDICTED: DNA repair protein RAD50 [Jatropha curcas] |
| 4 |
Hb_000813_100 |
0.0617956016 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000107_040 |
0.0666547296 |
- |
- |
nuclear pore complex protein nup153, putative [Ricinus communis] |
| 6 |
Hb_000023_060 |
0.0686984806 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 7 |
Hb_034243_020 |
0.0706133807 |
- |
- |
PREDICTED: elongator complex protein 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004543_030 |
0.0707905336 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 9 |
Hb_029142_010 |
0.074118047 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 10 |
Hb_001493_040 |
0.0742776212 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 11 |
Hb_004659_080 |
0.0770176438 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 12 |
Hb_001080_270 |
0.0774018339 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 13 |
Hb_000594_020 |
0.0782914012 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000227_380 |
0.0783686887 |
- |
- |
PREDICTED: protein TIME FOR COFFEE isoform X3 [Jatropha curcas] |
| 15 |
Hb_000120_610 |
0.0783731785 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 16 |
Hb_002471_210 |
0.0785818247 |
- |
- |
PREDICTED: THO complex subunit 2 [Jatropha curcas] |
| 17 |
Hb_056196_050 |
0.078844339 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 4-like [Jatropha curcas] |
| 18 |
Hb_000363_130 |
0.0791298981 |
- |
- |
PREDICTED: FRIGIDA-like protein 1 [Jatropha curcas] |
| 19 |
Hb_006252_020 |
0.0792472016 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 20 |
Hb_147737_010 |
0.0795369843 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |