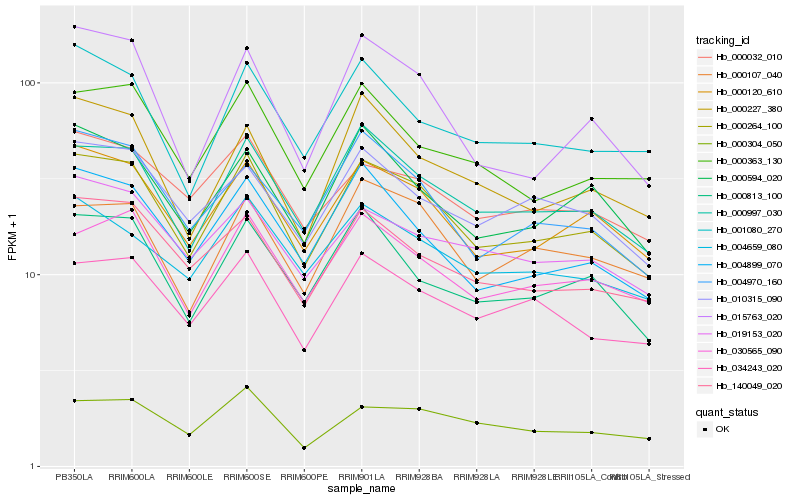
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000264_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000120_610 |
0.048414288 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 3 |
Hb_000997_030 |
0.0574265919 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 4 |
Hb_030565_090 |
0.0705719314 |
- |
- |
PREDICTED: DNA repair protein RAD50 [Jatropha curcas] |
| 5 |
Hb_140049_020 |
0.0747115177 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 6 |
Hb_000594_020 |
0.0753305908 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 7 |
Hb_004899_070 |
0.0761955101 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 8 |
Hb_034243_020 |
0.0764313772 |
- |
- |
PREDICTED: elongator complex protein 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000813_100 |
0.0764546572 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 10 |
Hb_004970_160 |
0.0777834627 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000304_050 |
0.0793992354 |
transcription factor |
TF Family: GRAS |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 12 |
Hb_000363_130 |
0.0798061039 |
- |
- |
PREDICTED: FRIGIDA-like protein 1 [Jatropha curcas] |
| 13 |
Hb_004659_080 |
0.080438273 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 14 |
Hb_000032_010 |
0.0807458377 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 15 |
Hb_000107_040 |
0.0808432133 |
- |
- |
nuclear pore complex protein nup153, putative [Ricinus communis] |
| 16 |
Hb_019153_020 |
0.0816737932 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000227_380 |
0.0828778951 |
- |
- |
PREDICTED: protein TIME FOR COFFEE isoform X3 [Jatropha curcas] |
| 18 |
Hb_015763_020 |
0.0831505413 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 19 |
Hb_001080_270 |
0.0834233842 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 20 |
Hb_010315_090 |
0.0835482064 |
- |
- |
suppressor of ty, putative [Ricinus communis] |