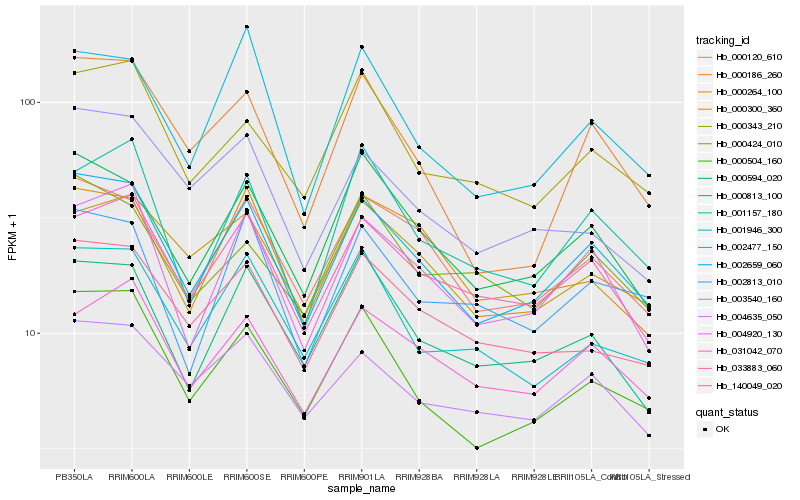
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002659_060 |
0.0 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 2 |
Hb_000120_610 |
0.0595052689 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 3 |
Hb_000594_020 |
0.0633578515 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000504_160 |
0.0642784832 |
- |
- |
hypothetical protein POPTR_0001s35340g [Populus trichocarpa] |
| 5 |
Hb_004920_130 |
0.0669413133 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_033883_060 |
0.0702800315 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 7 |
Hb_004635_050 |
0.0785591276 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 8 |
Hb_031042_070 |
0.0792814249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_140049_020 |
0.0802020957 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 10 |
Hb_000343_210 |
0.0809722157 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 11 |
Hb_000813_100 |
0.0826406807 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002477_150 |
0.0835164999 |
transcription factor |
TF Family: Alfin-like |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_001157_180 |
0.08599024 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_000300_360 |
0.0861102836 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 15 |
Hb_001946_300 |
0.0865692997 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 16 |
Hb_000264_100 |
0.087290779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000186_260 |
0.0878950141 |
transcription factor |
TF Family: PHD |
PREDICTED: protein polybromo-1-like [Jatropha curcas] |
| 18 |
Hb_002813_010 |
0.0894715262 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000424_010 |
0.0896270575 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003540_160 |
0.0899326419 |
- |
- |
hypothetical protein POPTR_0011s12160g [Populus trichocarpa] |