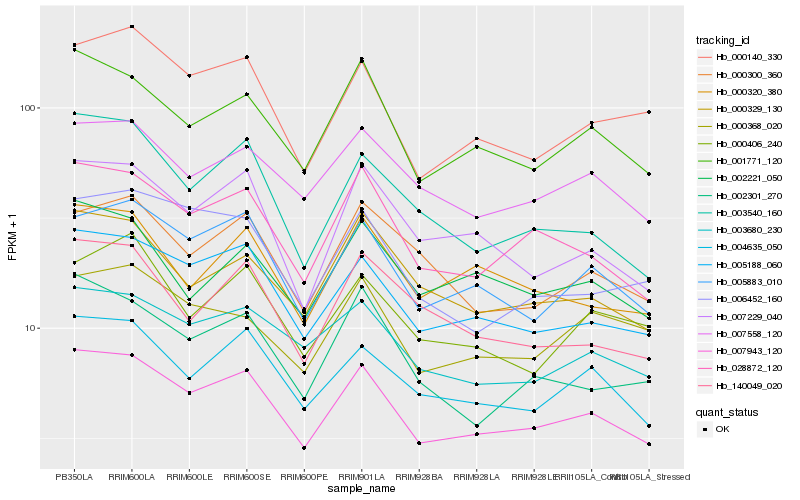
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007943_120 |
0.0 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 2 |
Hb_005188_060 |
0.0465275445 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 3 |
Hb_028872_120 |
0.0544343552 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 4 |
Hb_005883_010 |
0.0546342981 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001771_120 |
0.0573769557 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 6 |
Hb_004635_050 |
0.0605076141 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 7 |
Hb_003680_230 |
0.068516814 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 8 |
Hb_007558_120 |
0.0717776729 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 9 |
Hb_007229_040 |
0.0723921902 |
- |
- |
PREDICTED: protein ARABIDILLO 1-like [Jatropha curcas] |
| 10 |
Hb_000140_330 |
0.0728714097 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 11 |
Hb_000329_130 |
0.0748043553 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 12 |
Hb_003540_160 |
0.0784101864 |
- |
- |
hypothetical protein POPTR_0011s12160g [Populus trichocarpa] |
| 13 |
Hb_000320_380 |
0.0792614084 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002221_050 |
0.0814697011 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP65 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002301_270 |
0.0821514664 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 16 |
Hb_006452_160 |
0.0825328836 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 17 |
Hb_000300_360 |
0.0832947853 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 18 |
Hb_000368_020 |
0.0836937219 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_140049_020 |
0.0837452182 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 20 |
Hb_000406_240 |
0.0852019192 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |