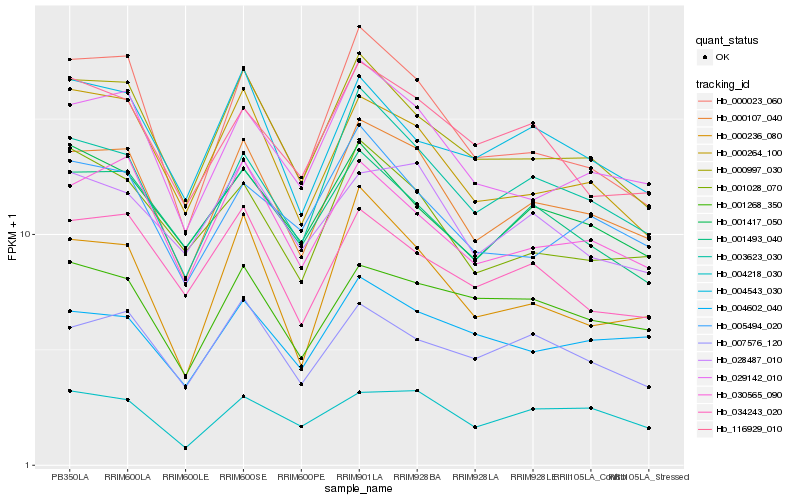
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_040 |
0.0 |
- |
- |
nuclear pore complex protein nup153, putative [Ricinus communis] |
| 2 |
Hb_000997_030 |
0.0666547296 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 3 |
Hb_029142_010 |
0.0678546323 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 4 |
Hb_003623_030 |
0.0693406687 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 14 [Jatropha curcas] |
| 5 |
Hb_000023_060 |
0.0761467282 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 6 |
Hb_030565_090 |
0.0782630598 |
- |
- |
PREDICTED: DNA repair protein RAD50 [Jatropha curcas] |
| 7 |
Hb_000264_100 |
0.0808432133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000236_080 |
0.0844006962 |
- |
- |
PREDICTED: small subunit processome component 20 homolog [Jatropha curcas] |
| 9 |
Hb_007576_120 |
0.0879166775 |
- |
- |
NBS-LRR disease resistance protein NBS49 [Dimocarpus longan] |
| 10 |
Hb_028487_010 |
0.0889892896 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001493_040 |
0.0891385684 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 12 |
Hb_116929_010 |
0.0892130671 |
- |
- |
PREDICTED: nuclear-pore anchor isoform X2 [Jatropha curcas] |
| 13 |
Hb_034243_020 |
0.0904435131 |
- |
- |
PREDICTED: elongator complex protein 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004218_030 |
0.0905055027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001028_070 |
0.0915789325 |
- |
- |
PREDICTED: WD repeat-containing protein 75 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005494_020 |
0.0926922904 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 17 |
Hb_004543_030 |
0.0934580268 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 18 |
Hb_004602_040 |
0.0950318453 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_001268_350 |
0.0951461116 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105115846 [Populus euphratica] |
| 20 |
Hb_001417_050 |
0.0957813701 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |