| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
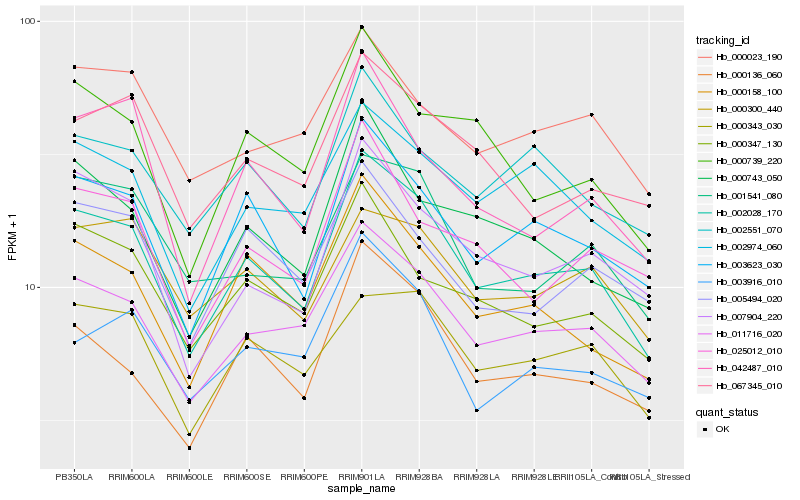
Hb_002028_170 |
0.0 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |
| 2 |
Hb_011716_020 |
0.0618208763 |
- |
- |
PREDICTED: uncharacterized protein LOC105637783 [Jatropha curcas] |
| 3 |
Hb_003623_030 |
0.0686093575 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 14 [Jatropha curcas] |
| 4 |
Hb_000158_100 |
0.0809930593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000136_060 |
0.0837153252 |
- |
- |
hypothetical protein POPTR_0016s06250g [Populus trichocarpa] |
| 6 |
Hb_007904_220 |
0.0881383224 |
- |
- |
PREDICTED: protein SDA1 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_000343_030 |
0.0883469091 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 8 |
Hb_002974_060 |
0.088692183 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X2 [Jatropha curcas] |
| 9 |
Hb_000347_130 |
0.0902969595 |
- |
- |
PREDICTED: nuclear pore complex protein NUP107 [Jatropha curcas] |
| 10 |
Hb_000743_050 |
0.0905453756 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_002551_070 |
0.092340856 |
- |
- |
hypothetical protein POPTR_0019s10130g [Populus trichocarpa] |
| 12 |
Hb_001541_080 |
0.092654896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000739_220 |
0.0938147111 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 14 |
Hb_067345_010 |
0.0948550137 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 15 |
Hb_042487_010 |
0.0955770952 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 9-like [Jatropha curcas] |
| 16 |
Hb_025012_010 |
0.0980621422 |
- |
- |
PREDICTED: importin-4 [Vitis vinifera] |
| 17 |
Hb_000300_440 |
0.0980834413 |
- |
- |
MIF4G domain and MA3 domain-containing protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_003916_010 |
0.0984861351 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 19 |
Hb_005494_020 |
0.0994490115 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 20 |
Hb_000023_190 |
0.0997398814 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |