| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
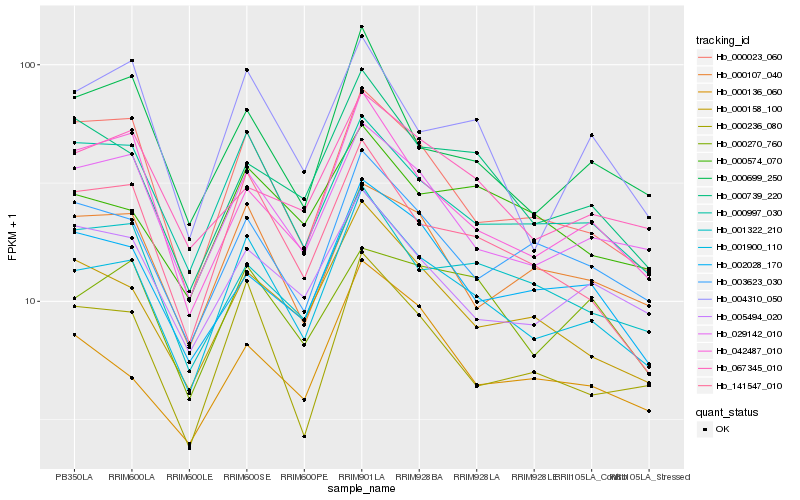
Hb_001900_110 |
0.0 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 2 |
Hb_141547_010 |
0.0843049473 |
- |
- |
PREDICTED: ARF guanine-nucleotide exchange factor GNOM [Jatropha curcas] |
| 3 |
Hb_000158_100 |
0.0852941651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_029142_010 |
0.0864321024 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 5 |
Hb_000997_030 |
0.0904621777 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 6 |
Hb_000023_060 |
0.0915789843 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 7 |
Hb_000236_080 |
0.0933219411 |
- |
- |
PREDICTED: small subunit processome component 20 homolog [Jatropha curcas] |
| 8 |
Hb_003623_030 |
0.0947032646 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 14 [Jatropha curcas] |
| 9 |
Hb_000739_220 |
0.097586113 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 10 |
Hb_042487_010 |
0.1012352294 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 9-like [Jatropha curcas] |
| 11 |
Hb_000699_250 |
0.1020083947 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 12 |
Hb_000107_040 |
0.1020818248 |
- |
- |
nuclear pore complex protein nup153, putative [Ricinus communis] |
| 13 |
Hb_004310_050 |
0.1049229104 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 14 |
Hb_005494_020 |
0.1063903714 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 15 |
Hb_000574_070 |
0.106401303 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 35 [Jatropha curcas] |
| 16 |
Hb_000136_060 |
0.1074261155 |
- |
- |
hypothetical protein POPTR_0016s06250g [Populus trichocarpa] |
| 17 |
Hb_002028_170 |
0.1077009027 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |
| 18 |
Hb_001322_210 |
0.110487823 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6-like [Jatropha curcas] |
| 19 |
Hb_000270_760 |
0.1124448222 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_067345_010 |
0.1131017937 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |