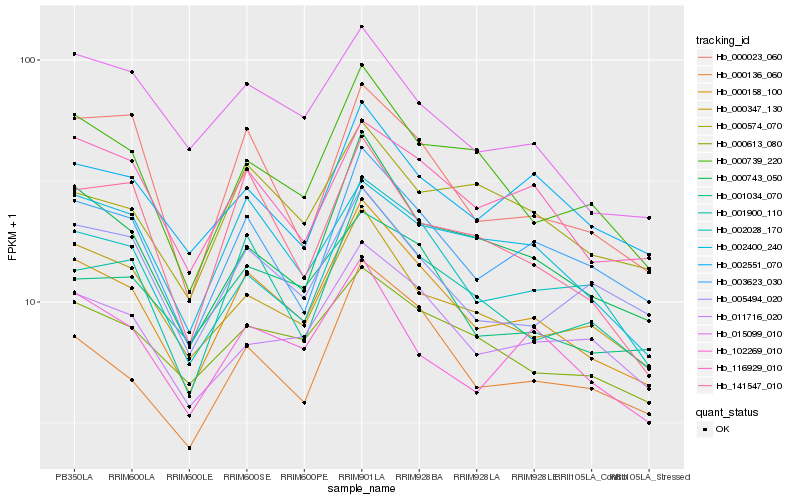
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000158_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000743_050 |
0.0761835716 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_003623_030 |
0.0769380558 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 14 [Jatropha curcas] |
| 4 |
Hb_000136_060 |
0.0797182072 |
- |
- |
hypothetical protein POPTR_0016s06250g [Populus trichocarpa] |
| 5 |
Hb_002028_170 |
0.0809930593 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |
| 6 |
Hb_000739_220 |
0.0847575478 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 7 |
Hb_001900_110 |
0.0852941651 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 8 |
Hb_116929_010 |
0.0884223107 |
- |
- |
PREDICTED: nuclear-pore anchor isoform X2 [Jatropha curcas] |
| 9 |
Hb_000347_130 |
0.0884592123 |
- |
- |
PREDICTED: nuclear pore complex protein NUP107 [Jatropha curcas] |
| 10 |
Hb_011716_020 |
0.0899833253 |
- |
- |
PREDICTED: uncharacterized protein LOC105637783 [Jatropha curcas] |
| 11 |
Hb_001034_070 |
0.090306585 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_000613_080 |
0.091224842 |
- |
- |
PREDICTED: mucin-5AC-like [Nicotiana sylvestris] |
| 13 |
Hb_102269_010 |
0.0942068471 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |
| 14 |
Hb_002551_070 |
0.0964080934 |
- |
- |
hypothetical protein POPTR_0019s10130g [Populus trichocarpa] |
| 15 |
Hb_000023_060 |
0.098807346 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 16 |
Hb_015099_010 |
0.0999654613 |
- |
- |
PREDICTED: probable glucan 1,3-alpha-glucosidase [Jatropha curcas] |
| 17 |
Hb_000574_070 |
0.1003327934 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 35 [Jatropha curcas] |
| 18 |
Hb_002400_240 |
0.1006924148 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 19 |
Hb_005494_020 |
0.1012559747 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 20 |
Hb_141547_010 |
0.1019016321 |
- |
- |
PREDICTED: ARF guanine-nucleotide exchange factor GNOM [Jatropha curcas] |