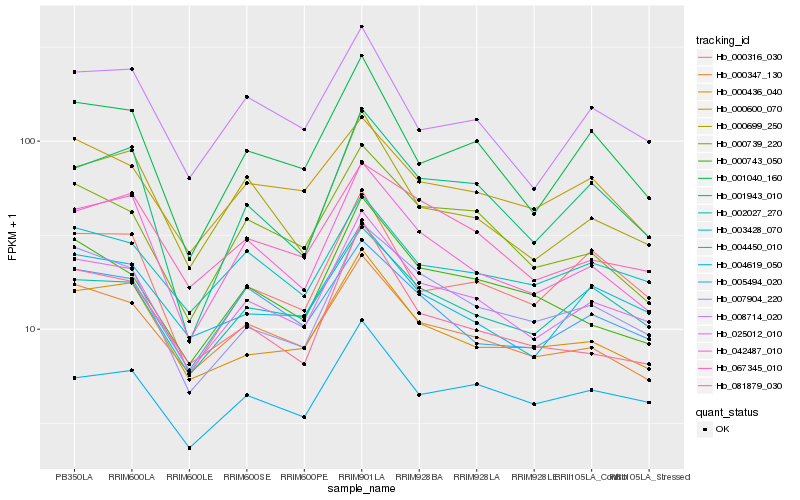
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025012_010 |
0.0 |
- |
- |
PREDICTED: importin-4 [Vitis vinifera] |
| 2 |
Hb_002027_270 |
0.0605591877 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL1 [Jatropha curcas] |
| 3 |
Hb_004619_050 |
0.0690189789 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 8-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000347_130 |
0.0691973855 |
- |
- |
PREDICTED: nuclear pore complex protein NUP107 [Jatropha curcas] |
| 5 |
Hb_007904_220 |
0.0716350162 |
- |
- |
PREDICTED: protein SDA1 homolog isoform X1 [Jatropha curcas] |
| 6 |
Hb_003428_070 |
0.0744228357 |
- |
- |
DEAD-box ATP-dependent RNA helicase 46 -like protein [Gossypium arboreum] |
| 7 |
Hb_001040_160 |
0.0758507156 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 8 |
Hb_008714_020 |
0.0763726623 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Jatropha curcas] |
| 9 |
Hb_000436_040 |
0.0776891472 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 10 |
Hb_004450_010 |
0.078253098 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 11 |
Hb_042487_010 |
0.0786440988 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 9-like [Jatropha curcas] |
| 12 |
Hb_000316_030 |
0.0788542262 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_067345_010 |
0.0790022304 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 14 |
Hb_005494_020 |
0.0798170009 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 15 |
Hb_000739_220 |
0.0800765354 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 16 |
Hb_000600_070 |
0.0830308473 |
- |
- |
PREDICTED: leucine aminopeptidase 1-like [Jatropha curcas] |
| 17 |
Hb_081879_030 |
0.0837348292 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 21 homolog [Jatropha curcas] |
| 18 |
Hb_001943_010 |
0.0840396049 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 19 |
Hb_000699_250 |
0.0843741425 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 20 |
Hb_000743_050 |
0.0862263814 |
- |
- |
unnamed protein product [Vitis vinifera] |