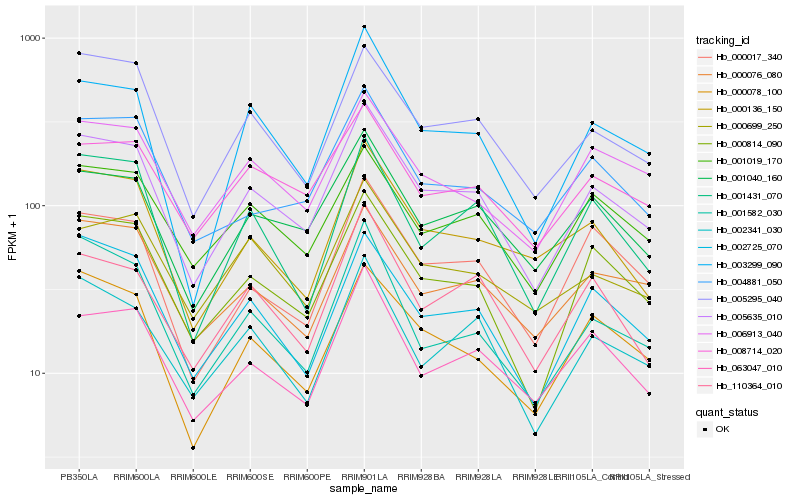
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005635_010 |
0.0 |
- |
- |
PREDICTED: cleft lip and palate transmembrane protein 1 homolog [Jatropha curcas] |
| 2 |
Hb_000814_090 |
0.0658443424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005295_040 |
0.0731754227 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP47-like [Jatropha curcas] |
| 4 |
Hb_001040_160 |
0.0737081937 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 5 |
Hb_002725_070 |
0.0786678256 |
- |
- |
hypothetical protein PRUPE_ppa005949mg [Prunus persica] |
| 6 |
Hb_003299_090 |
0.0808625818 |
- |
- |
hypothetical protein PRUPE_ppa011647mg [Prunus persica] |
| 7 |
Hb_000078_100 |
0.0811470335 |
- |
- |
PREDICTED: topless-related protein 4-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_063047_010 |
0.0842226558 |
- |
- |
peroxisomal targeting signal type 1 (pts1) receptor, putative [Ricinus communis] |
| 9 |
Hb_002341_030 |
0.0856576437 |
- |
- |
PREDICTED: transcription factor bHLH140 [Jatropha curcas] |
| 10 |
Hb_000017_340 |
0.0871436995 |
- |
- |
PREDICTED: GMP synthase [glutamine-hydrolyzing]-like [Phoenix dactylifera] |
| 11 |
Hb_000076_080 |
0.0875585342 |
- |
- |
PREDICTED: protein MOS2 [Jatropha curcas] |
| 12 |
Hb_001019_170 |
0.0883839946 |
- |
- |
PREDICTED: uncharacterized protein LOC105639322 [Jatropha curcas] |
| 13 |
Hb_008714_020 |
0.0889640774 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Jatropha curcas] |
| 14 |
Hb_006913_040 |
0.0895800119 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 15 |
Hb_110364_010 |
0.0899801105 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 16 |
Hb_001582_030 |
0.0904639622 |
- |
- |
PREDICTED: uncharacterized protein LOC105639462 [Jatropha curcas] |
| 17 |
Hb_004881_050 |
0.0905203856 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 18 |
Hb_000136_150 |
0.091394642 |
- |
- |
hypothetical protein VITISV_038366 [Vitis vinifera] |
| 19 |
Hb_000699_250 |
0.0942381388 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 20 |
Hb_001431_070 |
0.0953178098 |
- |
- |
PREDICTED: large proline-rich protein bag6-like isoform X2 [Jatropha curcas] |