| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
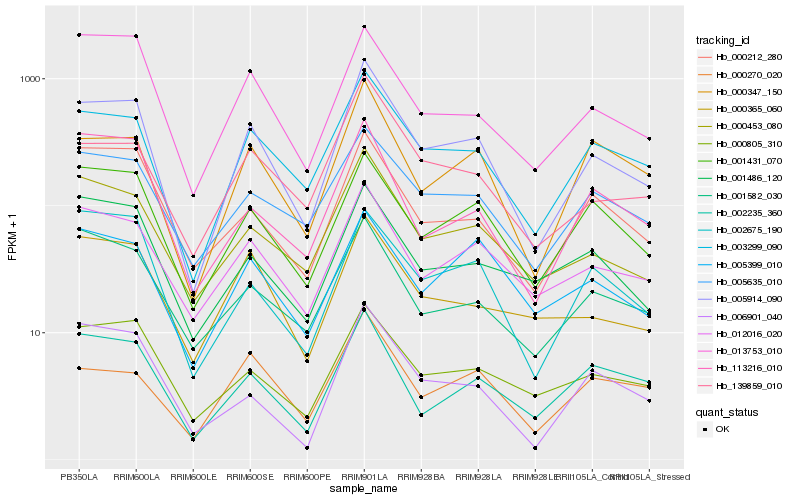
Hb_005914_090 |
0.0 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 2 |
Hb_003299_090 |
0.091910515 |
- |
- |
hypothetical protein PRUPE_ppa011647mg [Prunus persica] |
| 3 |
Hb_000453_080 |
0.1149389306 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_012016_020 |
0.1215138121 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105636265 [Jatropha curcas] |
| 5 |
Hb_000347_150 |
0.1222059461 |
- |
- |
Nucleolysin TIAR [Morus notabilis] |
| 6 |
Hb_001431_070 |
0.1231991856 |
- |
- |
PREDICTED: large proline-rich protein bag6-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_139859_010 |
0.1241769708 |
- |
- |
PREDICTED: cell division control protein 48 homolog D-like [Nicotiana sylvestris] |
| 8 |
Hb_013753_010 |
0.1256683779 |
- |
- |
PREDICTED: uncharacterized protein LOC105647969 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000805_310 |
0.1289445526 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005635_010 |
0.1293817666 |
- |
- |
PREDICTED: cleft lip and palate transmembrane protein 1 homolog [Jatropha curcas] |
| 11 |
Hb_000212_280 |
0.1336442494 |
- |
- |
PREDICTED: putative RNA-binding protein Luc7-like 2 [Gossypium raimondii] |
| 12 |
Hb_000365_060 |
0.1353856251 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |
| 13 |
Hb_002235_360 |
0.1357970191 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39680 [Jatropha curcas] |
| 14 |
Hb_002675_190 |
0.1383568243 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_001582_030 |
0.1384637056 |
- |
- |
PREDICTED: uncharacterized protein LOC105639462 [Jatropha curcas] |
| 16 |
Hb_006901_040 |
0.1395473384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005399_010 |
0.1423453126 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 18 |
Hb_113216_010 |
0.1438634808 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26-like [Jatropha curcas] |
| 19 |
Hb_001486_120 |
0.1441631668 |
- |
- |
PREDICTED: importin-5-like [Jatropha curcas] |
| 20 |
Hb_000270_020 |
0.1443320398 |
- |
- |
hypothetical protein POPTR_0015s07630g [Populus trichocarpa] |