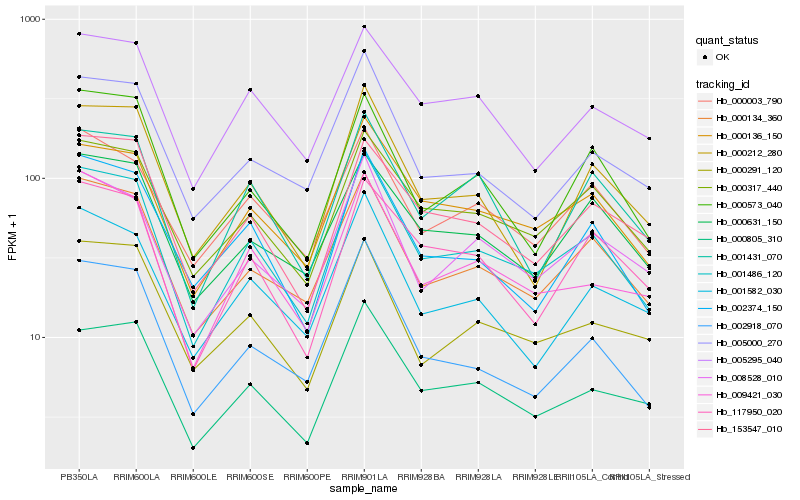
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001486_120 |
0.0 |
- |
- |
PREDICTED: importin-5-like [Jatropha curcas] |
| 2 |
Hb_000136_150 |
0.0652138681 |
- |
- |
hypothetical protein VITISV_038366 [Vitis vinifera] |
| 3 |
Hb_000134_360 |
0.0658742296 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 4 |
Hb_000805_310 |
0.0752774544 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000317_440 |
0.0771302579 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 13 isoform X2 [Jatropha curcas] |
| 6 |
Hb_009421_030 |
0.0792576177 |
- |
- |
PREDICTED: uncharacterized protein LOC105639700 [Jatropha curcas] |
| 7 |
Hb_008528_010 |
0.0823453008 |
- |
- |
PREDICTED: uncharacterized protein LOC105628498 [Jatropha curcas] |
| 8 |
Hb_002374_150 |
0.0826015535 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 9 |
Hb_002918_070 |
0.0840611532 |
- |
- |
hypothetical protein POPTR_0011s05530g [Populus trichocarpa] |
| 10 |
Hb_000212_280 |
0.088757096 |
- |
- |
PREDICTED: putative RNA-binding protein Luc7-like 2 [Gossypium raimondii] |
| 11 |
Hb_005000_270 |
0.0896341186 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_153547_010 |
0.0908008764 |
- |
- |
PREDICTED: protein FLX-like 4 [Jatropha curcas] |
| 13 |
Hb_000003_790 |
0.0913812244 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 4-like [Jatropha curcas] |
| 14 |
Hb_005295_040 |
0.0925757627 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP47-like [Jatropha curcas] |
| 15 |
Hb_001582_030 |
0.0932755511 |
- |
- |
PREDICTED: uncharacterized protein LOC105639462 [Jatropha curcas] |
| 16 |
Hb_117950_020 |
0.093719425 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Jatropha curcas] |
| 17 |
Hb_000631_150 |
0.0944991137 |
- |
- |
PREDICTED: RNA polymerase-associated protein LEO1 [Jatropha curcas] |
| 18 |
Hb_000573_040 |
0.0954517865 |
- |
- |
hypothetical protein JCGZ_19524 [Jatropha curcas] |
| 19 |
Hb_000291_120 |
0.0965589521 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 17 [Jatropha curcas] |
| 20 |
Hb_001431_070 |
0.0967547311 |
- |
- |
PREDICTED: large proline-rich protein bag6-like isoform X2 [Jatropha curcas] |