| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
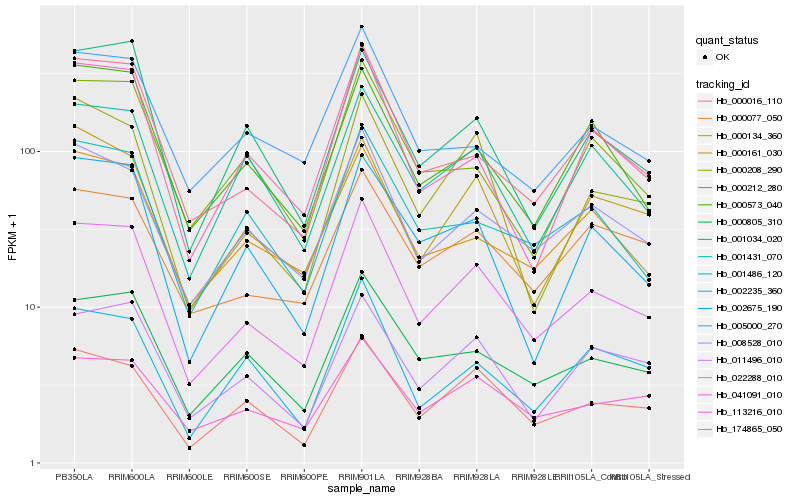
Hb_022288_010 |
0.0 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 2 |
Hb_008528_010 |
0.0836039888 |
- |
- |
PREDICTED: uncharacterized protein LOC105628498 [Jatropha curcas] |
| 3 |
Hb_000016_110 |
0.0884036268 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g38010 [Jatropha curcas] |
| 4 |
Hb_000805_310 |
0.0887731686 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic [Jatropha curcas] |
| 5 |
Hb_174865_050 |
0.0918635577 |
- |
- |
PREDICTED: neuroguidin [Jatropha curcas] |
| 6 |
Hb_011496_010 |
0.095956352 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g38420, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000208_290 |
0.1031023193 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_041091_010 |
0.1031579557 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001486_120 |
0.1061828147 |
- |
- |
PREDICTED: importin-5-like [Jatropha curcas] |
| 10 |
Hb_001431_070 |
0.107271101 |
- |
- |
PREDICTED: large proline-rich protein bag6-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000134_360 |
0.1072918737 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 12 |
Hb_113216_010 |
0.1092199338 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26-like [Jatropha curcas] |
| 13 |
Hb_002675_190 |
0.1096218406 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_000161_030 |
0.1104614487 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP8 [Jatropha curcas] |
| 15 |
Hb_000212_280 |
0.1105424078 |
- |
- |
PREDICTED: putative RNA-binding protein Luc7-like 2 [Gossypium raimondii] |
| 16 |
Hb_000077_050 |
0.1113046268 |
- |
- |
PREDICTED: elongation factor G-1, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_002235_360 |
0.1116681121 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39680 [Jatropha curcas] |
| 18 |
Hb_000573_040 |
0.1119972655 |
- |
- |
hypothetical protein JCGZ_19524 [Jatropha curcas] |
| 19 |
Hb_005000_270 |
0.1130916687 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_001034_020 |
0.1132058426 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |