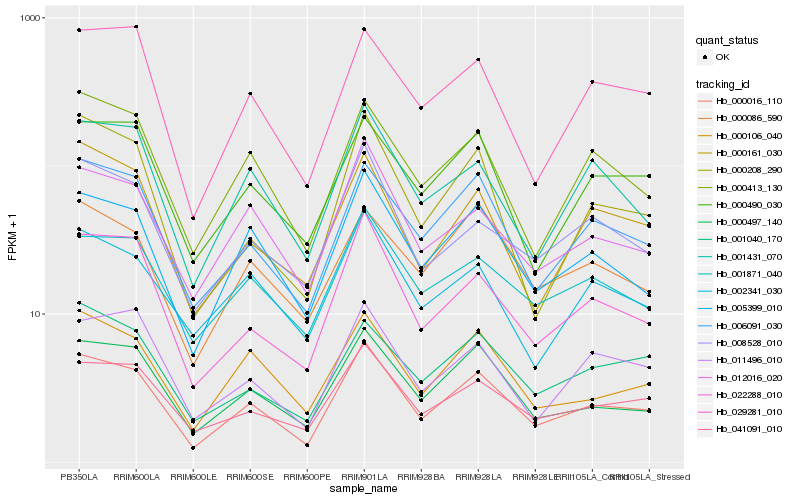
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000016_110 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g38010 [Jatropha curcas] |
| 2 |
Hb_005399_010 |
0.0784962736 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 3 |
Hb_000497_140 |
0.0808482978 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 4 |
Hb_006091_030 |
0.0820017271 |
desease resistance |
Gene Name: AAA |
PREDICTED: cell division control protein 48 homolog C-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_022288_010 |
0.0884036268 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 6 |
Hb_041091_010 |
0.0919490126 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 7 |
Hb_012016_020 |
0.0933873691 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105636265 [Jatropha curcas] |
| 8 |
Hb_000106_040 |
0.0958841549 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Vitis vinifera] |
| 9 |
Hb_000413_130 |
0.0979042743 |
- |
- |
PREDICTED: uncharacterized protein LOC105649409 [Jatropha curcas] |
| 10 |
Hb_001871_040 |
0.1015269453 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 1-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_001431_070 |
0.1025256462 |
- |
- |
PREDICTED: large proline-rich protein bag6-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_008528_010 |
0.1026668979 |
- |
- |
PREDICTED: uncharacterized protein LOC105628498 [Jatropha curcas] |
| 13 |
Hb_029281_010 |
0.1040291601 |
- |
- |
hypothetical protein JCGZ_04025 [Jatropha curcas] |
| 14 |
Hb_000490_030 |
0.1054692145 |
- |
- |
DNA-directed RNA polymerase II subunit, putative [Ricinus communis] |
| 15 |
Hb_000161_030 |
0.1058716233 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP8 [Jatropha curcas] |
| 16 |
Hb_011496_010 |
0.106762655 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g38420, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000208_290 |
0.1077254633 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001040_170 |
0.1109732601 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_002341_030 |
0.1110197304 |
- |
- |
PREDICTED: transcription factor bHLH140 [Jatropha curcas] |
| 20 |
Hb_000086_590 |
0.1129599949 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |