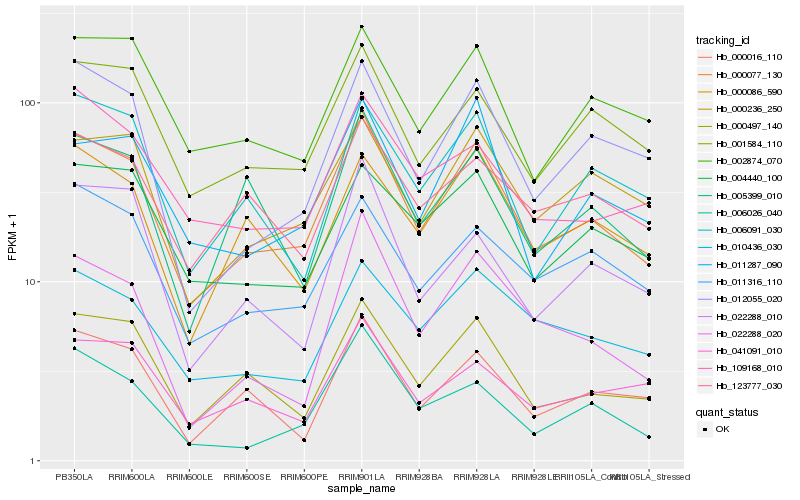
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_130 |
0.0 |
- |
- |
PREDICTED: callose synthase 12 [Jatropha curcas] |
| 2 |
Hb_000497_140 |
0.0891663775 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 3 |
Hb_001584_110 |
0.105330032 |
- |
- |
PREDICTED: importin subunit alpha-4 isoform X2 [Jatropha curcas] |
| 4 |
Hb_012055_020 |
0.1068501396 |
- |
- |
PREDICTED: metal transporter Nramp3 [Jatropha curcas] |
| 5 |
Hb_000236_250 |
0.1095043313 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002874_070 |
0.1114815016 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |
| 7 |
Hb_006091_030 |
0.1115170279 |
desease resistance |
Gene Name: AAA |
PREDICTED: cell division control protein 48 homolog C-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_011316_110 |
0.1116850809 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At4g25390 [Jatropha curcas] |
| 9 |
Hb_010436_030 |
0.1143052007 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 10 |
Hb_000016_110 |
0.1144823195 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g38010 [Jatropha curcas] |
| 11 |
Hb_004440_100 |
0.115029448 |
- |
- |
PREDICTED: transmembrane protein 53 [Jatropha curcas] |
| 12 |
Hb_000086_590 |
0.1159382493 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 13 |
Hb_011287_090 |
0.116124259 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 14 |
Hb_005399_010 |
0.1189907378 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 15 |
Hb_109168_010 |
0.1197327039 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 16 |
Hb_041091_010 |
0.1197378443 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 17 |
Hb_006026_040 |
0.1199718569 |
- |
- |
PREDICTED: 60S ribosomal protein L13a-4-like [Jatropha curcas] |
| 18 |
Hb_123777_030 |
0.1200308086 |
transcription factor |
TF Family: SNF2 |
PREDICTED: DNA helicase INO80 isoform X2 [Jatropha curcas] |
| 19 |
Hb_022288_020 |
0.121770391 |
- |
- |
hypothetical protein POPTR_0006s08590g [Populus trichocarpa] |
| 20 |
Hb_022288_010 |
0.1236141206 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |