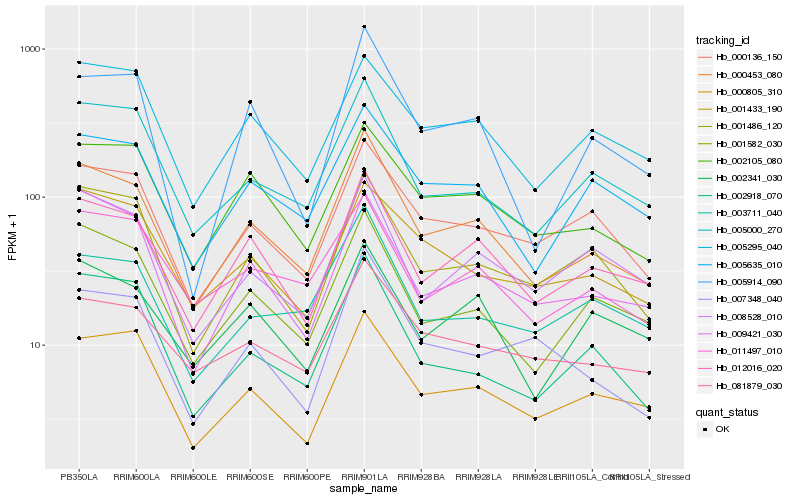
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000453_080 |
0.0 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_012016_020 |
0.0795193568 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105636265 [Jatropha curcas] |
| 3 |
Hb_001582_030 |
0.0912338117 |
- |
- |
PREDICTED: uncharacterized protein LOC105639462 [Jatropha curcas] |
| 4 |
Hb_005000_270 |
0.0972698729 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_005635_010 |
0.0985631465 |
- |
- |
PREDICTED: cleft lip and palate transmembrane protein 1 homolog [Jatropha curcas] |
| 6 |
Hb_002105_080 |
0.10012987 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 7 |
Hb_001486_120 |
0.1036203406 |
- |
- |
PREDICTED: importin-5-like [Jatropha curcas] |
| 8 |
Hb_000805_310 |
0.1041229808 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000136_150 |
0.10456102 |
- |
- |
hypothetical protein VITISV_038366 [Vitis vinifera] |
| 10 |
Hb_009421_030 |
0.1078153443 |
- |
- |
PREDICTED: uncharacterized protein LOC105639700 [Jatropha curcas] |
| 11 |
Hb_011497_010 |
0.10814496 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 12 |
Hb_002918_070 |
0.1101952342 |
- |
- |
hypothetical protein POPTR_0011s05530g [Populus trichocarpa] |
| 13 |
Hb_005295_040 |
0.1109718801 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP47-like [Jatropha curcas] |
| 14 |
Hb_081879_030 |
0.1113554987 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 21 homolog [Jatropha curcas] |
| 15 |
Hb_003711_040 |
0.1128520415 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 16 |
Hb_008528_010 |
0.1130898058 |
- |
- |
PREDICTED: uncharacterized protein LOC105628498 [Jatropha curcas] |
| 17 |
Hb_005914_090 |
0.1149389306 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 18 |
Hb_002341_030 |
0.1169644315 |
- |
- |
PREDICTED: transcription factor bHLH140 [Jatropha curcas] |
| 19 |
Hb_001433_190 |
0.1188924583 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit A [Jatropha curcas] |
| 20 |
Hb_007348_040 |
0.119077419 |
transcription factor |
TF Family: PHD |
hypothetical protein RCOM_1314010 [Ricinus communis] |