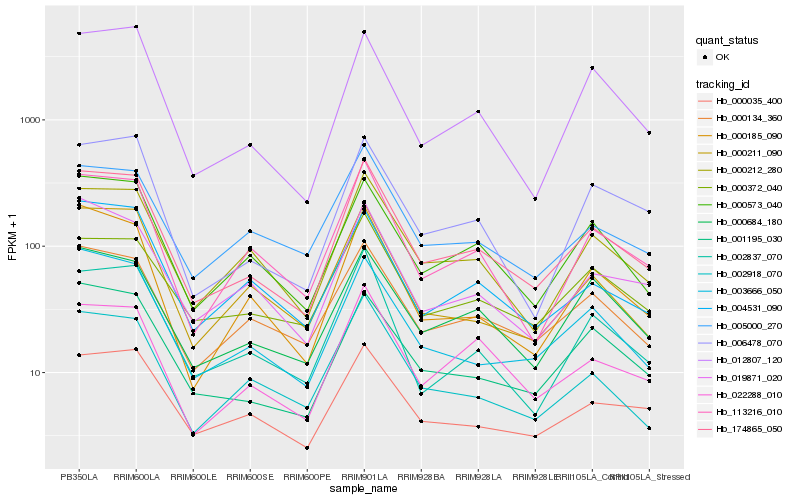
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_174865_050 |
0.0 |
- |
- |
PREDICTED: neuroguidin [Jatropha curcas] |
| 2 |
Hb_000212_280 |
0.0807286277 |
- |
- |
PREDICTED: putative RNA-binding protein Luc7-like 2 [Gossypium raimondii] |
| 3 |
Hb_000573_040 |
0.0842034628 |
- |
- |
hypothetical protein JCGZ_19524 [Jatropha curcas] |
| 4 |
Hb_113216_010 |
0.0845249849 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26-like [Jatropha curcas] |
| 5 |
Hb_003666_050 |
0.0855209797 |
- |
- |
PREDICTED: NASP-related protein sim3 [Jatropha curcas] |
| 6 |
Hb_001195_030 |
0.0863024046 |
- |
- |
auxilin, putative [Ricinus communis] |
| 7 |
Hb_000185_090 |
0.0890051064 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 8 |
Hb_002837_070 |
0.0895027879 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 8-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_022288_010 |
0.0918635577 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 10 |
Hb_000134_360 |
0.0919405214 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 11 |
Hb_005000_270 |
0.0930756026 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_019871_020 |
0.0931922313 |
- |
- |
PREDICTED: uncharacterized protein LOC105642047 [Jatropha curcas] |
| 13 |
Hb_002918_070 |
0.093337109 |
- |
- |
hypothetical protein POPTR_0011s05530g [Populus trichocarpa] |
| 14 |
Hb_000211_090 |
0.0953992496 |
- |
- |
hypothetical protein PHAVU_011G023400g, partial [Phaseolus vulgaris] |
| 15 |
Hb_000684_180 |
0.0962761837 |
- |
- |
PREDICTED: importin subunit alpha-9 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004531_090 |
0.0983035726 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 17 |
Hb_006478_070 |
0.0986342421 |
- |
- |
PREDICTED: uncharacterized protein LOC105649957 [Jatropha curcas] |
| 18 |
Hb_012807_120 |
0.0986689661 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_000035_400 |
0.0992080227 |
- |
- |
PREDICTED: lariat debranching enzyme-like [Citrus sinensis] |
| 20 |
Hb_000372_040 |
0.100793262 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |