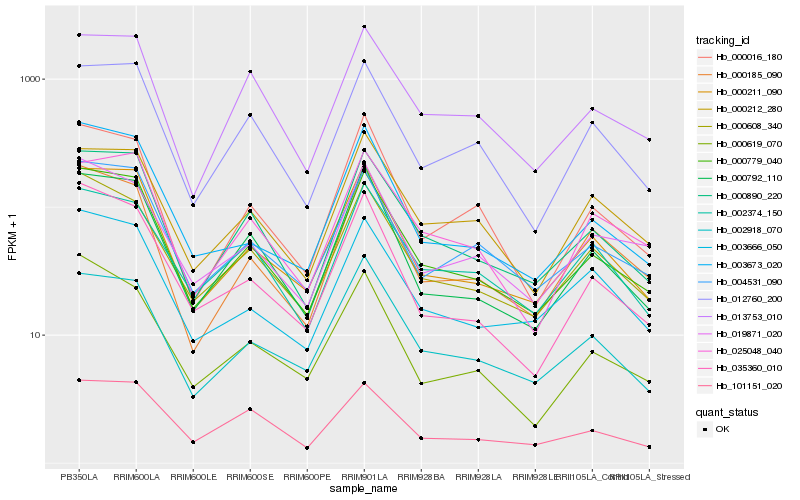
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000779_040 |
0.0 |
- |
- |
PREDICTED: protein FLX-like 4 [Jatropha curcas] |
| 2 |
Hb_000890_220 |
0.0437112679 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative chromatin-remodeling complex ATPase chain isoform X1 [Jatropha curcas] |
| 3 |
Hb_000211_090 |
0.0548801064 |
- |
- |
hypothetical protein PHAVU_011G023400g, partial [Phaseolus vulgaris] |
| 4 |
Hb_000608_340 |
0.0687726218 |
- |
- |
PREDICTED: putative RNA-binding protein Luc7-like 2 [Populus euphratica] |
| 5 |
Hb_000792_110 |
0.0697172461 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 6 |
Hb_004531_090 |
0.0699493769 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 7 |
Hb_003673_020 |
0.0796003929 |
- |
- |
hypothetical protein JCGZ_20998 [Jatropha curcas] |
| 8 |
Hb_035360_010 |
0.0840324195 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 9 |
Hb_002918_070 |
0.0850393126 |
- |
- |
hypothetical protein POPTR_0011s05530g [Populus trichocarpa] |
| 10 |
Hb_000185_090 |
0.086243537 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 11 |
Hb_101151_020 |
0.0865794564 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_003666_050 |
0.0871490821 |
- |
- |
PREDICTED: NASP-related protein sim3 [Jatropha curcas] |
| 13 |
Hb_012760_200 |
0.0879604816 |
- |
- |
protein phsophatase-2a, putative [Ricinus communis] |
| 14 |
Hb_013753_010 |
0.0905492523 |
- |
- |
PREDICTED: uncharacterized protein LOC105647969 isoform X1 [Jatropha curcas] |
| 15 |
Hb_019871_020 |
0.0905544991 |
- |
- |
PREDICTED: uncharacterized protein LOC105642047 [Jatropha curcas] |
| 16 |
Hb_000016_180 |
0.0929902299 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26-like [Jatropha curcas] |
| 17 |
Hb_025048_040 |
0.0932286264 |
- |
- |
PREDICTED: NAP1-related protein 2 [Jatropha curcas] |
| 18 |
Hb_002374_150 |
0.093607279 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 19 |
Hb_000212_280 |
0.0944172991 |
- |
- |
PREDICTED: putative RNA-binding protein Luc7-like 2 [Gossypium raimondii] |
| 20 |
Hb_000619_070 |
0.0955486164 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH1 [Jatropha curcas] |