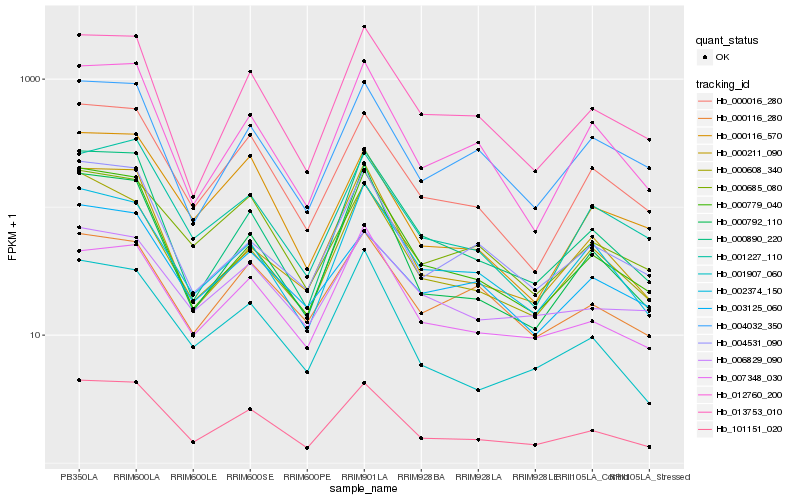
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_101151_020 |
0.0 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000016_280 |
0.0734866779 |
- |
- |
Arginine/serine-rich splicing factor 35 [Theobroma cacao] |
| 3 |
Hb_000890_220 |
0.0767547581 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative chromatin-remodeling complex ATPase chain isoform X1 [Jatropha curcas] |
| 4 |
Hb_013753_010 |
0.0840758175 |
- |
- |
PREDICTED: uncharacterized protein LOC105647969 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000779_040 |
0.0865794564 |
- |
- |
PREDICTED: protein FLX-like 4 [Jatropha curcas] |
| 6 |
Hb_007348_030 |
0.0865944575 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X2 [Jatropha curcas] |
| 7 |
Hb_012760_200 |
0.0908829848 |
- |
- |
protein phsophatase-2a, putative [Ricinus communis] |
| 8 |
Hb_002374_150 |
0.0911921643 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 9 |
Hb_001907_060 |
0.092554573 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 5 [Jatropha curcas] |
| 10 |
Hb_001227_110 |
0.0953764483 |
- |
- |
U2 snrnp auxiliary factor, small subunit, putative [Ricinus communis] |
| 11 |
Hb_004531_090 |
0.095706033 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 12 |
Hb_000792_110 |
0.0965900212 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 13 |
Hb_004032_350 |
0.0977248639 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like isoform X1 [Musa acuminata subsp. malaccensis] |
| 14 |
Hb_000608_340 |
0.0990409843 |
- |
- |
PREDICTED: putative RNA-binding protein Luc7-like 2 [Populus euphratica] |
| 15 |
Hb_000685_080 |
0.0994713256 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_006829_090 |
0.100844783 |
- |
- |
PREDICTED: protein MOS2 [Jatropha curcas] |
| 17 |
Hb_000211_090 |
0.1028986698 |
- |
- |
hypothetical protein PHAVU_011G023400g, partial [Phaseolus vulgaris] |
| 18 |
Hb_000116_570 |
0.1039026181 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 19 |
Hb_003125_060 |
0.1043744985 |
- |
- |
PREDICTED: snurportin-1 [Jatropha curcas] |
| 20 |
Hb_000116_280 |
0.1044886515 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |