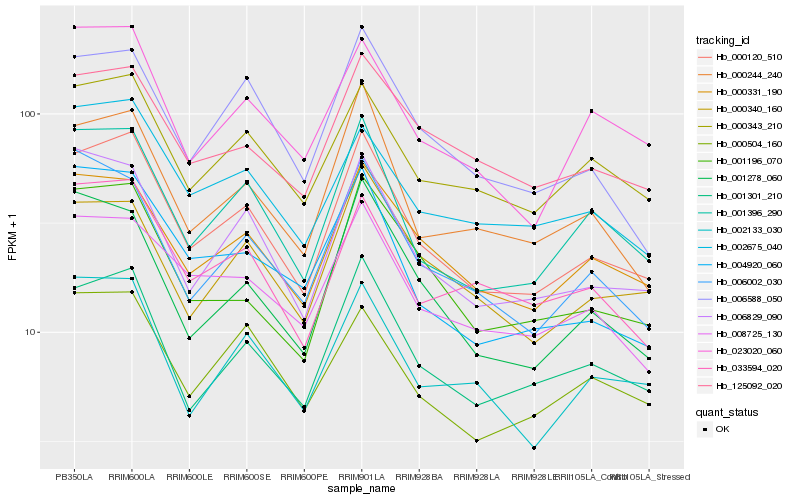
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_510 |
0.0 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001396_290 |
0.0627782833 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |
| 3 |
Hb_001301_210 |
0.0632563262 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 32 [Jatropha curcas] |
| 4 |
Hb_006829_090 |
0.0687081102 |
- |
- |
PREDICTED: protein MOS2 [Jatropha curcas] |
| 5 |
Hb_000331_190 |
0.074122964 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 6 |
Hb_000343_210 |
0.0763174298 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 7 |
Hb_000244_240 |
0.0782616599 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_002675_040 |
0.0834801249 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 9 |
Hb_008725_130 |
0.0838831311 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 10 |
Hb_001196_070 |
0.0850269604 |
- |
- |
nucleolar protein nop56, putative [Ricinus communis] |
| 11 |
Hb_023020_060 |
0.0857550112 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 12 |
Hb_000504_160 |
0.0859872846 |
- |
- |
hypothetical protein POPTR_0001s35340g [Populus trichocarpa] |
| 13 |
Hb_002133_030 |
0.0863759891 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 1 [Jatropha curcas] |
| 14 |
Hb_001278_060 |
0.0867687694 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 15 |
Hb_006588_050 |
0.0868343933 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 16 |
Hb_033594_020 |
0.0874590353 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 17 |
Hb_006002_030 |
0.0884748004 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 18 |
Hb_125092_020 |
0.088622378 |
- |
- |
PREDICTED: nucleolin 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000340_160 |
0.089134197 |
- |
- |
PREDICTED: elongation factor Tu, mitochondrial [Jatropha curcas] |
| 20 |
Hb_004920_060 |
0.0893539616 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |