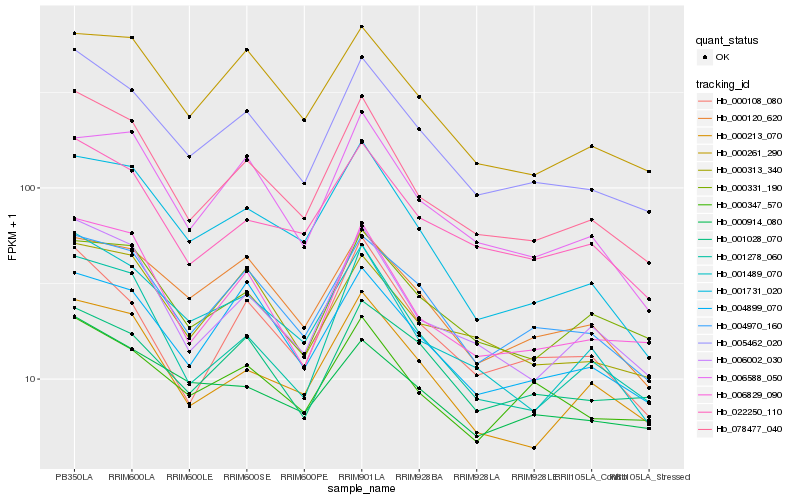
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005462_020 |
0.0 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 2 |
Hb_078477_040 |
0.0569343446 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |
| 3 |
Hb_004970_160 |
0.0668945068 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_006829_090 |
0.0775631551 |
- |
- |
PREDICTED: protein MOS2 [Jatropha curcas] |
| 5 |
Hb_006002_030 |
0.0796211064 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 6 |
Hb_022250_110 |
0.0806390778 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_000120_620 |
0.0823524678 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 8 |
Hb_004899_070 |
0.0827233572 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 9 |
Hb_000261_290 |
0.0852820462 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 10 |
Hb_000313_340 |
0.0856453234 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 11 |
Hb_001028_070 |
0.086071223 |
- |
- |
PREDICTED: WD repeat-containing protein 75 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001489_070 |
0.0862567972 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 13 |
Hb_000347_570 |
0.0881313663 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 14 |
Hb_000914_080 |
0.089415525 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_001731_020 |
0.0903044356 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 16 |
Hb_000108_080 |
0.0914072112 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 17 |
Hb_001278_060 |
0.0915991051 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 18 |
Hb_006588_050 |
0.0917439038 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 19 |
Hb_000213_070 |
0.0919207615 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor At2g41710 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000331_190 |
0.0922939277 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |