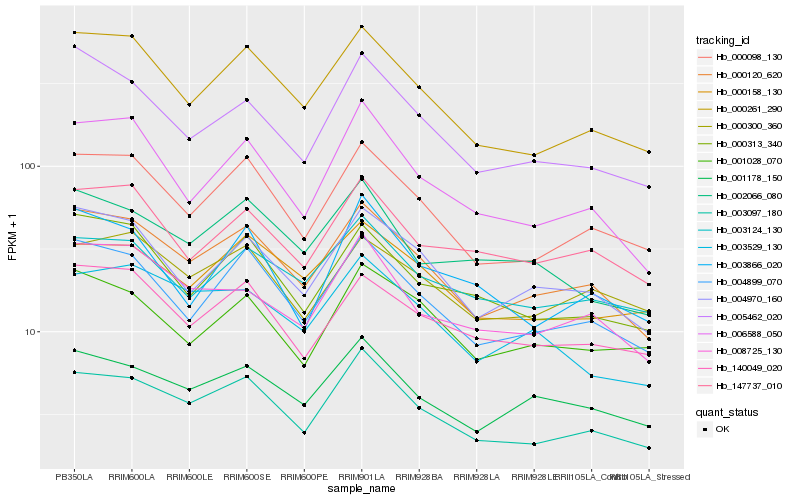
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003097_180 |
0.0 |
- |
- |
PREDICTED: crossover junction endonuclease MUS81 [Jatropha curcas] |
| 2 |
Hb_000120_620 |
0.0589180194 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 3 |
Hb_000098_130 |
0.0593126295 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 4 |
Hb_000261_290 |
0.0684034194 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 5 |
Hb_004899_070 |
0.0741647717 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 6 |
Hb_006588_050 |
0.0788478157 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 7 |
Hb_001178_150 |
0.0878467519 |
- |
- |
PREDICTED: uncharacterized protein LOC105635484 [Jatropha curcas] |
| 8 |
Hb_000158_130 |
0.0911415711 |
- |
- |
PREDICTED: uncharacterized protein LOC105647950 isoform X1 [Jatropha curcas] |
| 9 |
Hb_008725_130 |
0.0926404439 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 10 |
Hb_003124_130 |
0.0942669475 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003866_020 |
0.0958518079 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004970_160 |
0.096398914 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_002066_080 |
0.0966988596 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 14 |
Hb_005462_020 |
0.0971905058 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 15 |
Hb_003529_130 |
0.0975054911 |
- |
- |
PREDICTED: ribosome biogenesis protein BOP1 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_000313_340 |
0.0984922256 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 17 |
Hb_000300_360 |
0.0997297187 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 18 |
Hb_147737_010 |
0.1011174714 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_140049_020 |
0.1016942395 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 20 |
Hb_001028_070 |
0.1025715637 |
- |
- |
PREDICTED: WD repeat-containing protein 75 isoform X2 [Jatropha curcas] |